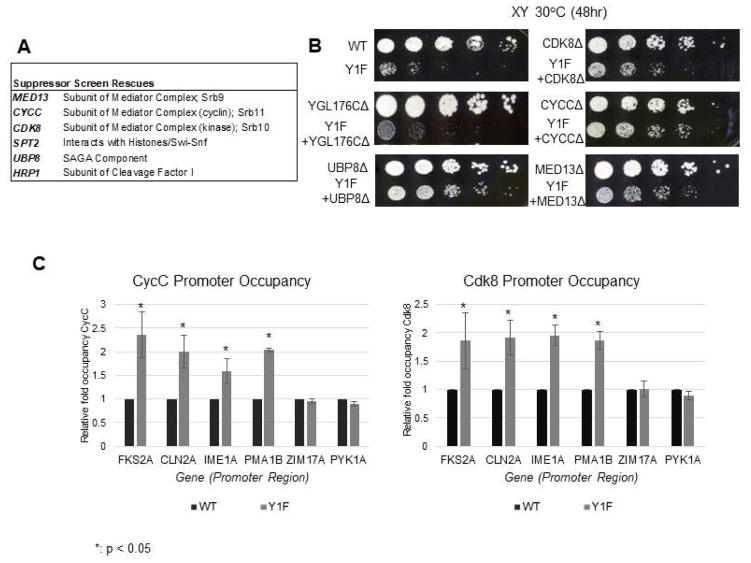
Figure 2. SGA analysis identifies suppressors of Y1F growth defects.
(A) List of genes showing synthetic rescue of Y1F strain in two SGA suppressor screens. Genes displaying strongest interactions and involved in RNAP II transcription are shown (refer to Table S2 for a complete list). (B) Confirmation of genetic interaction between Y1F and MED13/CDK8/CYCC and UBP8. Strains of indicated genotypes were generated by recombinant transformation and growth compared by spot assay on rich medium with serial five-fold dilutions. YGL178C showed no interaction with Y1F in SGA screen and serves as a control. (C) ChIP analysis of Cdk8/CycC occupancy at promoter regions of indicated genes, using 3HA-tagged Cdk8/CycC. Data normalized to WT signal at each gene; P values less than 0.05 are indicated (*). ChIP data is represented as mean +/− SE of three independent experiments.