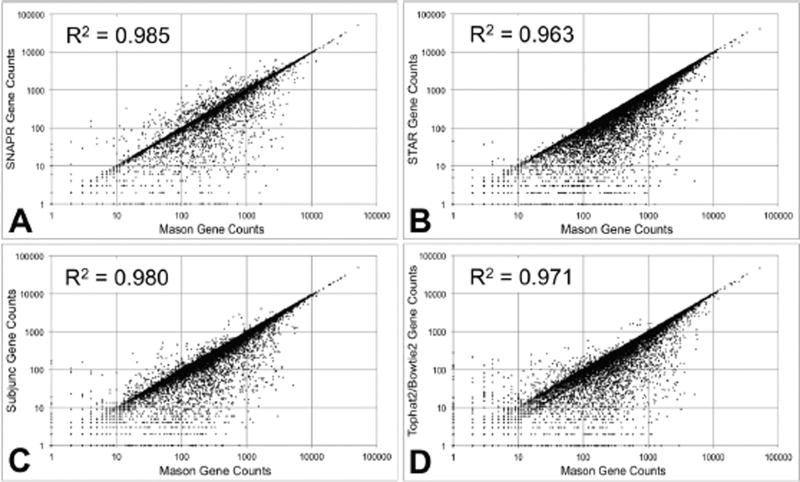
Figure 2.
Aligned gene read counts for 2×107 paired-end reads generated by Mason with Illumina™ sequencing errors using the Venter genome.100% of reads were generated from the Ensembl v68 annotation, and aligned using each of the four tested aligners. ‘Correct’ read counts (x-axis)for each gene were generated from the Mason SAM file. (A) SNAPR automatically generates gene read counts. (B) Read counts for STAR alignments were generated using htseq-count. (C) Read counts for Subjunc alignments were generated using the featureCounts functionality. (D) Read counts for Tophat2/Bowtie2 alignments were generated using htseq-count.