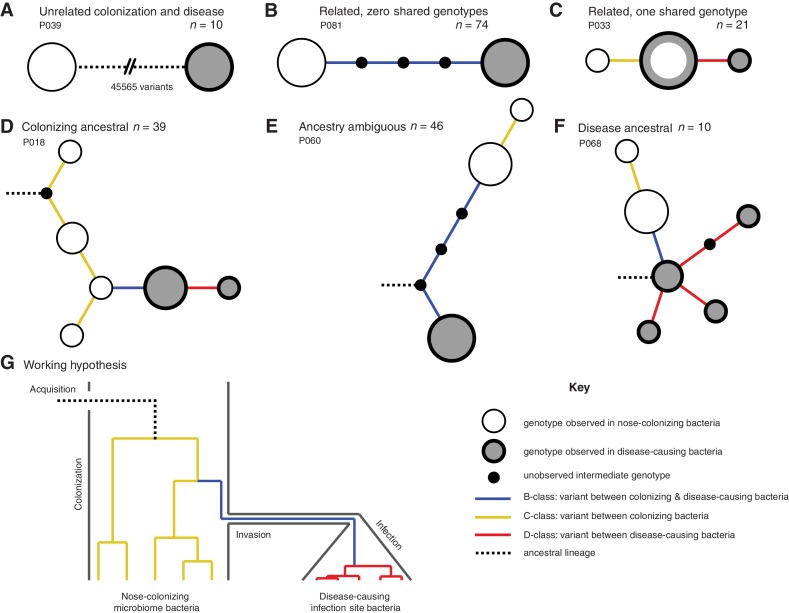
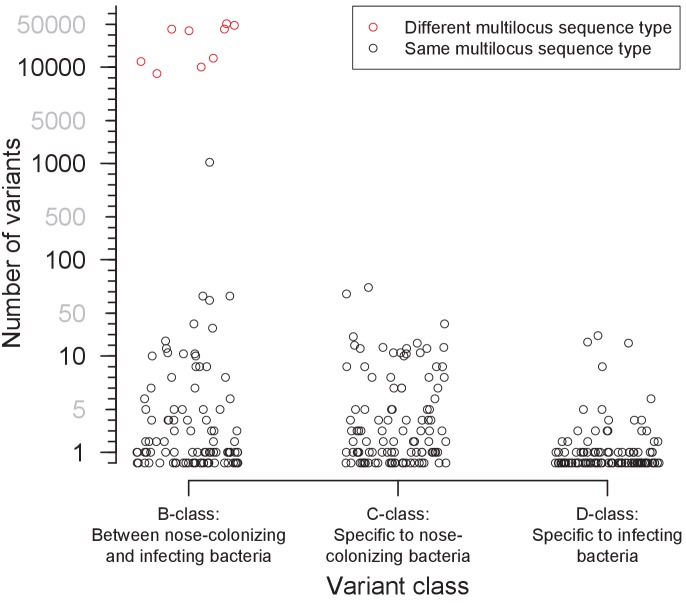
Figure 1. Infection-causing S. aureus form closely related but distinct populations descended from nose-colonizing bacteria in the majority of infections.
Bacteria sampled from the nose and infection site of 105 patients formed one of three population structures, illustrated with example haplotrees: (A) Unrelated populations differentiated by many variants. (B) Highly related populations separated by few variants. (C) Highly related populations with one genotype in common. Reconstructing the ancestral genotype in each patient helped identify the ancestral population: (D) Nose-colonizing bacteria ancestral. (E) Ambiguous ancestral population. (F) Infection site bacteria ancestral. (G) Phylogeny illustrating the working hypothesis that variants differentiating highly related nose-colonizing and infection-causing bacteria would be enriched for variants that promote, or are promoted by, infection. In A–F, haplotree nodes represent observed genotypes sampled from the nose (white) or infection site (grey), with area proportional to genotype frequency, or unobserved intermediate genotypes (black). Edges represent mutations. Patient identifiers and sample sizes (n) are given. In A–G, edge color indicates that mutations occurring on those branches correspond to B-class variants between nose-colonizing and infection-causing bacteria (blue), C-class variants among nose-colonizing bacteria (gold) or D-class variants among infection-causing bacteria (red). Black dashed edges indicate ancestral lineages. A B C.