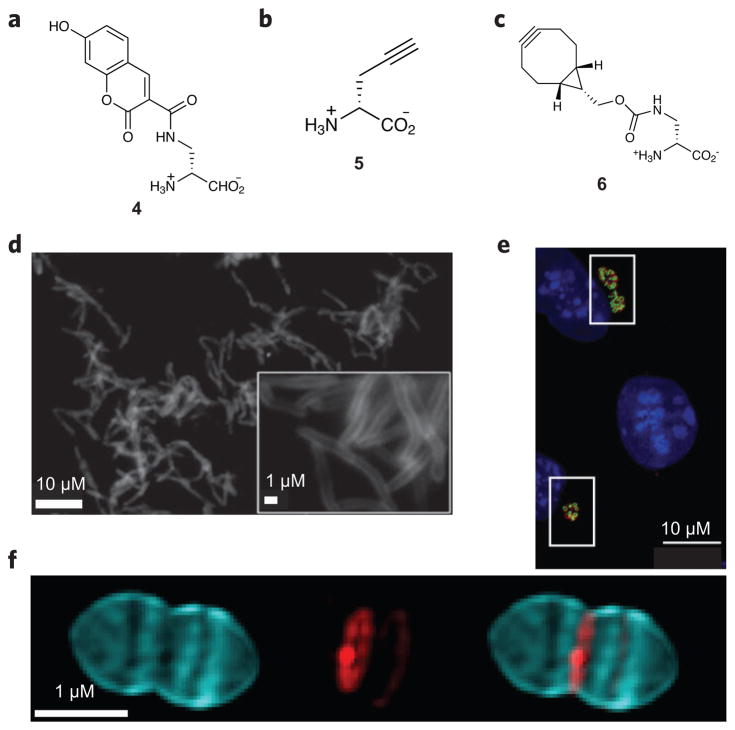
Figure 4. Peptidoglycan-inspired probes and images obtained with these compounds.
(a,b) Fluorescent D-amino acids hydroxycoumarin-carbonyl-amino-D-alanine (4; a) and (R)-propargylglycine (5; b) for tracking PG production. (c) The cyclooctyne D-alanine analog endobcnDala (6) used for no-wash PG labeling. (d) Fluorescence image of M. smegmatis treated with 6 and labeled with bis-sulfated glycol-substituted azido silicon rhodamine. (e) Fluorescence image of mouse fibroblast cells (L2 cells) infected with C. trachomatis and labeled with the dipeptide ethynyl-D-alanyl-D-alanine followed by an azide-modified Alexa Fluor 488 (green). An antibody for chlamydial major outer membrane protein (red) was used to label chlamydial elementary bodies and reticulate bodies, and 4′,6-diamidino-2-phenylindole (DAPI; blue) was used for nuclear staining. (f) 3D-SIM images of S. pneumoniae PG pulse-chase labeled with 4 (blue) and tetramethylrhodamine 3-amino-D-alanine (red). Using this strategy, older PG (blue) can be differentiated from areas of new PG synthesis (red). Figures reproduced with permission from: d, ref. 57; e, ref. 65; f, ref. 67.