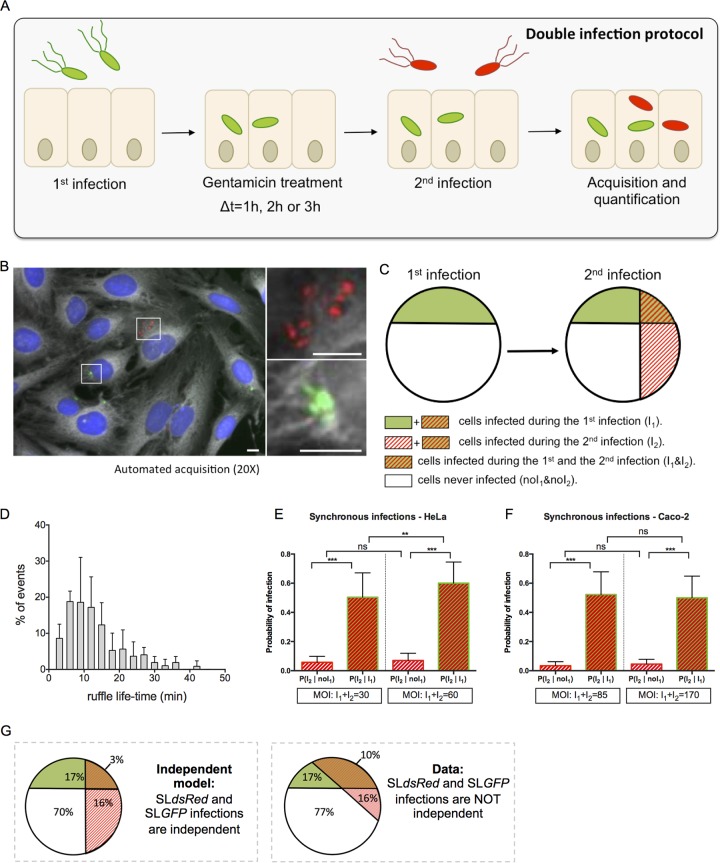
FIG 1.
Double infections allow studies of Salmonella cooperation at the single-cell level. (A) An overview of the experimental workflow used in this study is shown. According to the sequential infection protocol, HeLa cells grown in 96-well plates for 24 h were subjected for 30 min to a first infection by SLGFP. This was followed by elimination of extracellular bacteria by gentamicin and incubation of the cells for 1, 2, or 3 h. The cells were subsequently challenged by a second infection with SLdsRed for 30 min. After removal of the extracellular bacteria, the samples were fixed. Nuclei were stained with DAPI, and cell membranes were stained with CellMask before microscopic acquisition of the entire wells. (B) Representative image of SLGFP and SLdsRed internalized in HeLa cells. Host cell nuclei are visualized through DAPI staining (blue), and cell membranes are visualized with CellMask (gray). Scale bar, 5 μm. (C) Scheme of our statistical analysis of different subpopulations. The following cellular populations can be distinguished: those cells infected during the first infection (I1) or not (noI1), those infected during the second infection (I2) or not (noI2), and the related subpopulations (I1 & I2; noI1 & noI2). This scheme maps the case of two independent infections. (D) Time distribution of the ruffle disappearance during Salmonella infection followed in actin-GFP transfected cells by time-lapse microscopy. (E and F) Comparison of the conditional probabilities of infection for two different populations during synchronous infection of SLGFP and SLdsRed in HeLa cells and Caco-2 cells. The MOIs were chosen to obtain an average of 30% of the cells infected and calculated after CFU counting (n ≥ 3). P values were obtained after a t test. (G) Comparison of an independent model (left) with the obtained data (right). The percentages are averaged from six independent experiments, represented in panel E, at an MOI of 30. **, P < 0.01; ***, P < 0.001; ns, not significant.