FIG 4.
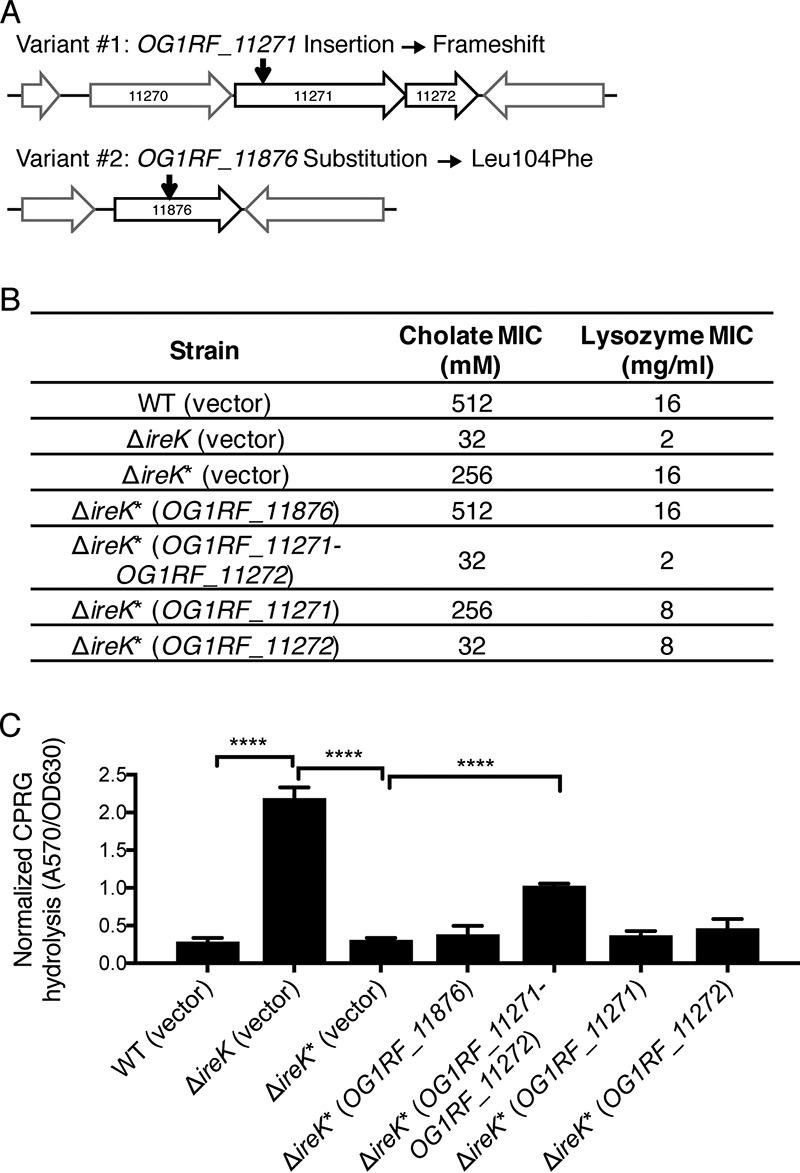
Disruption of OG1RF_11271 and OG1RF_11272 drives the phenotype of ΔireK* suppressor mutants. (A) Genetic architecture near the nucleotide variants uncovered by whole-genome sequencing (the drawing is not to scale). The locations of the nucleotide variants are indicated by black arrows. (B) The cholate and lysozyme resistance of the WT E. faecalis strain (OG1RF), the ΔireK mutant (CK119), and the ΔireK* suppressor mutant carrying the empty vector (pJRG9) or expressing wild-type copies of the indicated genes was determined. The reported MICs represent the median values from three independent biological replicates. (C) CPRG hydrolysis was measured for the strains listed in panel B. The reported measurements represent averages from three independent cultures, and error bars represent standard deviations. Statistical significance was evaluated by t test. ****, P < 0.0001.
