Abstract
Glioma is the most common malignant tumor in the central nervous system. This study aims to explore the potential mechanism and identify gene signatures of glioma. The glioma gene expression profile GSE4290 was analyzed for differentially expressed genes (DEGs). Gene ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) analyses were applied for the enriched pathways. A protein-protein interaction (PPI) network was constructed to find the hub genes. Survival analysis was conducted to screen and validate critical genes. In this study, 775 downregulated DEGs were identified. GO analysis demonstrated that the DEGs were enriched in cellular protein modification, regulation of cell communication, and regulation of signaling. KEGG analysis indicated that the DEGs were enriched in the MAPK signaling pathway, endocytosis, oxytocin signaling, and calcium signaling. PPI network and module analysis found 12 hub genes, which were enriched in synaptic vesicle cycling rheumatoid arthritis and collecting duct acid secretion. The four key genes CDK17, GNA13, PHF21A, and MTHFD2 were identified in both generation (GSE4412) and validation (GSE4271) dataset, respectively. Regression analysis showed that CDK13, PHF21A, and MTHFD2 were independent predictors. The results suggested that CDK17, GNA13, PHF21A, and MTHFD2 might play important roles and potentially be valuable in the prognosis and treatment of glioma.
1. Introduction
Among the various histological subtypes of brain tumor, glioma is the most common malignant tumor in the central nervous system [1]. Established by the World Health Organization (WHO), it can be classified from grade I to grade IV based on histopathological and clinical criteria [2]. During invasive growth, most gliomas extend processes, resulting in a lack of clear borders between tumor and normal brain tissue, making surgical resection of the entire carcinoma difficult. Currently, imageological examination is the most important diagnostic method, as well as the evaluation of the postoperative curative effect. However, imaging is influenced by many factors, such as radiation injury and surgery that result in poor specificity. It is difficult to achieve early diagnosis and treatment of glioma due to a lack of specificity of auxiliary examination indices, so that many patients can lose the chance for radical excision, thereby increasing the risk for poor prognosis. The 5-year overall survival (OS) of patients with glioblastoma is less than 10% [3]. Therefore, the identification of sensitive and specific biological markers that would help identify patients at a higher or lower risk of death from glioma is of vital importance, not only for a better understanding of the molecular and cellular processes involved in tumorigenesis but also for more effective diagnosis, suitable treatment, and improved prognosis.
Gene expression profiling analysis is a useful method with broad clinical application for identifying tumor-related genes in various types of cancer, from molecular diagnosis to pathological classification, from therapeutic evaluation to prognosis prediction, and from drug sensitivity to neoplasm recurrence [4–6]. However, the use of microarrays in clinical practice is limited by the overwhelming number of genes identified by gene profiling, lack of both repeatability and independent validation, and need for complicated statistical analyses [7]. Therefore, in order to put these expression profiles in clinical practice, it is necessary to identify a suitable number of genes and develop a method that can be operated by routine assay. In this study, we downloaded original data from the glioma microarray in the Gene Expression Omnibus (GEO, http://www.ncbi.nlm.nih.gov/geo/), an online public collection database for registration, which is not only for saving microarray data but also for helping the user query and download. We compare gene expression profiles of tumor cells with normal brain cells in order to identify differentially expressed genes (DEGs). Subsequently, the identified DEGs were screened by using Morpheus online software, followed by gene ontology (GO) and pathway enrichment analysis. After analyzing their biological functions and pathways, we further explored the potential biomarkers for diagnosis and prognosis by survival analysis in two independent datasets in order to gain insight on glioma development and progression at the molecular level.
2. Materials and Methods
2.1. Microarray Data
We downloaded the gene expression profiles in GSE4290, GSE4412, and GSE4271 from the GEO database. GSE4290 has a total of 180 samples, including 157 cases of glioma (26 astrocytomas, 50 oligodendrogliomas, and 81 glioblastomas) and 23 cases of normal brain tissue, based on the GPL570 platform (Affymetrix Human Genome U133 Plus 2.0 Array) by Fine HA et al. Using the GPL96 platform (Affymetrix Human Genome U133A Array), the GES4412 dataset containing 85 cases of glioma was submitted by Nelson SF; and the GES4271 dataset, containing 100 samples that included 77 cases of primary tumor samples and 23 cases of recurrence, was submitted by Phillips HS et al.
2.2. Screen Genes of Differential Expression
The analysis was carried out by using GEO2R, an online analysis tool, for the GEO database, based on R language. We applied analysis to classify the sample into two groups that had similar expression patterns in glioma and normal brain tissue. We defined DEGs as differentially expressed with logFC > 2 or logFC < −2, a criteria as described in the references [8, 9]. An adj. P value < 0.05 was considered statistically significant. In addition, we used visual hierarchical cluster analysis to show the two groups by Morpheus online analysis software (https://software.broadinstitute.org/morpheus/) after the relative raw data of TXT files was downloaded.
2.3. Gene Ontology and KEGG Pathway Analysis of DEGs
With functions including molecular function, biological pathways, and cellular component, gene ontology (GO) analysis annotates genes and gene products [10]. KEGG comprises a set of genome and enzymatic approaches and a biological chemical energy online database [11]. It is a resource for systematic analysis of gene function and related high-level genome functional information. DAVID (https://david.ncifcrf.gov/) can provide systematic and comprehensive biological function annotation information for high-throughput gene expression [12]. Therefore, we applied GO and KEGG pathway analyses to the DEGs by using DAVID online tools at the functional level. A P < 0.05 was considered to have significant differences.
2.4. Integration of Protein-Protein Interaction (PPI) Network and Module Analysis
The STRING database is an online tool for assessment and integration of protein-protein interactions, including direct (physical), as well as indirect (functional) associations. STRING version 10.0 covers 9,643,763 proteins from 2031 organisms [13]. We drew DEGs using STRING in order to assess the interactional relationships among the DEGs, then used the Cytoscape software to build a PPI network, employed the plug-in Molecular Complex Detection (MCODE) to screen PPI network modules, and established scores > 3 and nodes > 4 in the MCODE module, function, and pathway enrichment analysis. A P < 0.05 was considered statistically significant.
2.5. Identification of Biomarkers
Based on the information in the individual MCODE modules, the node with the highest score was selected as the hub gene in GSE4290. Every hub gene was also found in two independent datasets (generation dataset GSE4412, primary tumor samples n = 85, and validation dataset GSE4271, primary tumor samples n = 77) based on the downloaded raw data files, including the information of gene expression value, overall survival time (OS), and survival state. Statistical analyses were performed using SPSS version 20.0 for Windows (IBM, Chicago, IL). We divided expression values into two groups, high expression and low expression, according to X-tile [14]. The Kaplan-Meier method was used to determine the probability of survival and analyzed by the log-rank test. A P < 0.05 was considered statistically significant.
3. Results
3.1. Identification of DEGs
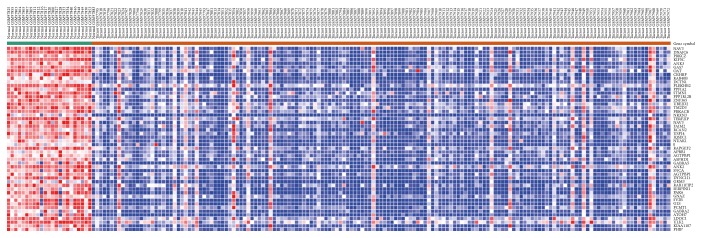
A comparison of 157 cases of glioma samples with 23 cases of normal brain tissue in GSE4290 by using the GEO2R online analysis tool resulted in the identification of the DEGs listed in Figure 1. Based on GEO2R analysis, using an adjusted P < 0.05 and log (fold change) (logFC) ≥ 2.0 criteria, there were 775 downregulated genes identified. We further validated the results by using the Morpheus online tool, resulting in a DEG expression heat map, of the top 50 downregulated genes, shown in Figure 1.
Figure 1.
Heat map of the top 50 downregulated genes (red: upregulated; purple: downregulated).
3.2. Gene Ontology Enrichment Analysis
We used the DAVID online analysis tool to identify statistically significantly enriched GO terms and KEGG pathways after uploading all downregulated genes. GO analysis results showed that downregulated DEGs were significantly enriched in molecular function (MF), including small molecule binding, nucleoside phosphate binding, and carbohydrate derivative binding (Table 1). For biological processes (BP), the downregulated genes were enriched in cellular protein modification, regulation of cell communication, and regulation of signaling (Table 1). In addition, GO cell component (CC) analysis also displayed that the downregulated DEGs were significantly enriched in the cytosol, membrane-bounded vesicles, and nucleoplasm (Table 1).
Table 1.
Gene ontology analysis of downregulated genes associated with glioma.
| Category | Term/gene function | Count | % | P value |
|---|---|---|---|---|
| GOTERM_MF_FAT | GO:0036094~small molecule binding | 119 | 0.125 | 2.76E − 06 |
| GOTERM_MF_FAT | GO:0000166~nucleotide binding | 116 | 0.121 | 3.45E − 07 |
| GOTERM_MF_FAT | GO:1901265~nucleoside phosphate binding | 116 | 0.121 | 3.52E − 07 |
| GOTERM_MF_FAT | GO:0097367~carbohydrate derivative binding | 100 | 0.105 | 1.00E − 04 |
| GOTERM_MF_FAT | GO:0019899~enzyme binding | 94 | 0.098 | 2.24E − 07 |
| GOTERM_MF_FAT | GO:0017076~purine nucleotide binding | 92 | 0.096 | 9.44E − 06 |
| GOTERM_MF_FAT | GO:0032555~purine ribonucleotide binding | 91 | 0.095 | 1.26E − 05 |
| GOTERM_MF_FAT | GO:0032553~ribonucleotide binding | 91 | 0.095 | 1.76E − 05 |
| GOTERM_BP_FAT | GO:0036211~protein modification process | 148 | 0.155 | 8.31E − 04 |
| GOTERM_BP_FAT | GO:0006464~cellular protein modification process | 148 | 0.155 | 8.31E − 04 |
| GOTERM_BP_FAT | GO:0023051~regulation of signaling | 140 | 0.147 | 3.36E − 07 |
| GOTERM_BP_FAT | GO:0010646~regulation of cell communication | 139 | 0.146 | 2.25E − 07 |
| GOTERM_BP_FAT | GO:0033036~macromolecule localization | 138 | 0.145 | 5.25E − 09 |
| GOTERM_BP_FAT | GO:0006793~phosphorus metabolic process | 134 | 0.140 | 1.05E − 05 |
| GOTERM_BP_FAT | GO:0006796~phosphate-containing compound metabolic process | 133 | 0.139 | 1.53E − 05 |
| GOTERM_BP_FAT | GO:0008104~protein localization | 124 | 0.130 | 6.40E − 09 |
| GOTERM_CC_FAT | GO:0005829~cytosol | 163 | 0.171 | 4.20E − 08 |
| GOTERM_CC_FAT | GO:0031988~membrane-bounded vesicle | 152 | 0.159 | 2.84E − 04 |
| GOTERM_CC_FAT | GO:0005654~nucleoplasm | 130 | 0.136 | 2.88E − 04 |
| GOTERM_CC_FAT | GO:0097458~neuron part | 91 | 0.095 | 1.38E − 11 |
| GOTERM_CC_FAT | GO:0030054~cell junction | 75 | 0.078 | 1.15E − 05 |
| GOTERM_CC_FAT | GO:0043005~neuron projection | 66 | 0.069 | 1.72E − 08 |
| GOTERM_CC_FAT | GO:0005794~Golgi apparatus | 66 | 0.069 | 0.008029 |
| GOTERM_CC_FAT | GO:0016023~cytoplasmic, membrane-bounded vesicle | 64 | 0.067 | 7.15E − 05 |
3.3. KEGG Pathway Analysis
The significant enriched pathways of the downregulated DEGs, analyzed by KEGG analysis, are shown in Table 2. The downregulated genes were enriched in the MAPK signaling pathway, endocytosis, oxytocin signaling pathway, calcium signaling pathway, proteoglycans in cancer, purine metabolism, cAMP signaling pathway, and regulation of the actin cytoskeleton.
Table 2.
KEGG pathway analysis of downregulation genes associated with glioma.
| Term | Pathway | Gene count | % | P value | Genes |
|---|---|---|---|---|---|
| hsa04010 | MAPK signaling pathway | 22 | 0.023 | 6.35E − 05 | MEF2C, BRAF, MAP2K1, NLK, MAP2K4, TP53, PPP3R1, PTPRR, CACNG3, PRKCG, PRKCB, CDC42, RASGRF2, MAPK9, MAPK8IP3, STMN1, PAK1, PRKACB, RAPGEF2, CACNA1C, DUSP7, CACNA1B |
| hsa04144 | Endocytosis | 20 | 0.021 | 5.98E − 04 | SH3GL3, PARD3, CLTB, PSD3, PIP5K1C, HLA-E, EPS15, RAB11FIP4, MVB12A, CDC42, AP2A2, RAB31, SH3GLB2, NEDD4, ARPC5L, ARF3, KIAA1033, NEDD4L, IQSEC1, F2R |
| hsa04921 | Oxytocin signaling pathway | 15 | 0.015 | 5.25E − 04 | MEF2C, ADCY2, CAMK1G, MAP2K1, PPP1R12B, PPP3R1, CACNG3, PRKCG, CAMKK1, PRKCB, CAMKK2, CAMK2B, GUCY1B3, PRKACB, CACNA1C |
| hsa04020 | Calcium signaling pathway | 14 | 0.014 | 0.0048 | SLC8A1, ADCY2, PTGER3, CCKBR, PPP3R1, PRKCG, PRKCB, ATP2B1,PDE1A, CAMK2B, PRKACB, CACNA1C, CACNA1B, F2R |
| hsa05205 | Proteoglycans in cancer | 14 | 0.014 | 0.012 | CDC42, WNT10B, HIF1A, MAP2K1, BRAF, ANK3, PPP1R12B, TP53, PRKCG, CAMK2B, PAK1, PRKACB, TIMP3, PRKCB |
| hsa00230 | Purine metabolism | 13 | 0.013 | 0.0109 | NME4, ADSS, GDA, ADCY2, POLR1E, RRM2, PDE1A, PRIM2, AK5, GUCY1B3, ENTPD4, HPRT1, GART |
| hsa04024 | cAMP signaling pathway | 13 | 0.013 | 0.0253 | PPARA, ADCY2, PTGER3, BRAF, MAP2K1, ATP2B1, NPY, MAPK9, CAMK2B, PRKACB, PAK1, CACNA1C, F2R |
| hsa04810 | Regulation of actin cytoskeleton | 13 | 0.013 | 0.0387 | GNA13, PAK6, CDC42, ENAH,MAP2K1, BRAF, ARHGEF6, PPP1R12B, ARPC5L, WASF2, PIP5K1C, PAK1, F2R |
3.4. Module Analysis and Hub Gene Selection in the PPI Network
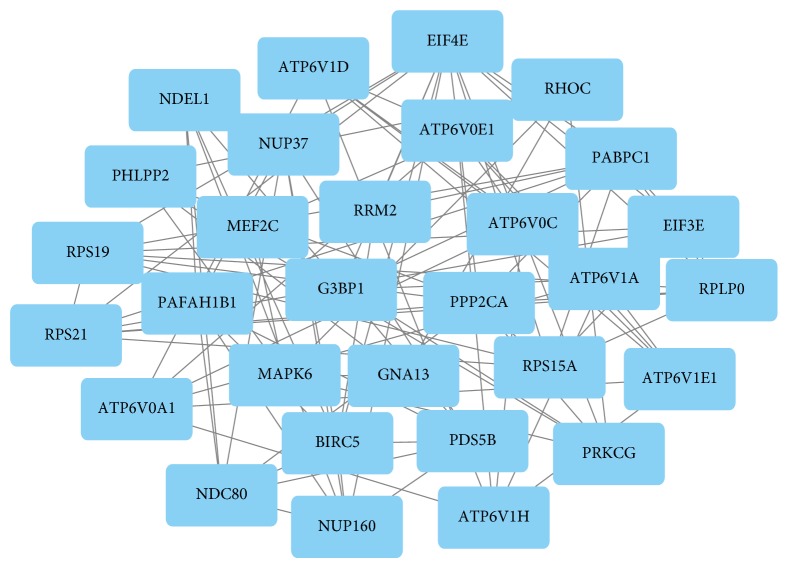
Based on the information in the STRING database, the highest module was shown by using the MCODE plug-in, and the functional annotation of the genes involved in the module was analyzed (Figure 2). Enrichment pathway analysis showed that the genes in the module were related to synaptic vesicle cycling, rheumatoid arthritis, and collecting duct acid secretion. Moreover, the 12 hub nodes of the highest score were screened in all modules. The hub genes included PDS5 cohesin associated factor B (PDS5B), chromodomain helicase DNA binding protein 5 (CHD5), cyclin-dependent kinase 17 (CDK17), eukaryotic translation initiation factor 3 subunit E (EIF3E), ATPase H+ transporting V1 subunit H (ATP6V1H), G protein subunit alpha 13 (GNA13), PHD finger protein 21A (PHF21A), methylenetetrahydrofolate dehydrogenase (NADP+ dependent) 2 (MTHFD2), lipoprotein lipase (LPL), adenylosuccinate synthase (ADSS), Wnt family member 10B (WNT10B), and serine and arginine-rich splicing factor 1 (SRSF1). The enriched pathways for genes in the highest module were shown in Table 3.
Figure 2.
Highest module selected from the PPI network.
Table 3.
The enriched pathways for genes in the highest module.
| Pathway | P value | FDR | Nodes |
|---|---|---|---|
| Synaptic vesicle cycle | 3.36E − 08 | 3.58E − 05 | ATP6V0C, ATP6V1A, ATP6V0E1, ATP6V1E1, ATP6V1H, ATP6V0A1, ATP6V1D |
| Rheumatoid arthritis | 3.43E − 07 | 3.66E − 04 | ATP6V0C, ATP6V1A, ATP6V0E1, ATP6V1E1, ATP6V1H, ATP6V0A1, ATP6V1D |
| Collecting duct acid secretion | 2.29E − 08 | 2.44E − 05 | ATP6V0C, ATP6V1A, ATP6V0E1, ATP6V1E1, ATP6V0A1, ATP6V1D |
3.5. Identification of Biomarkers
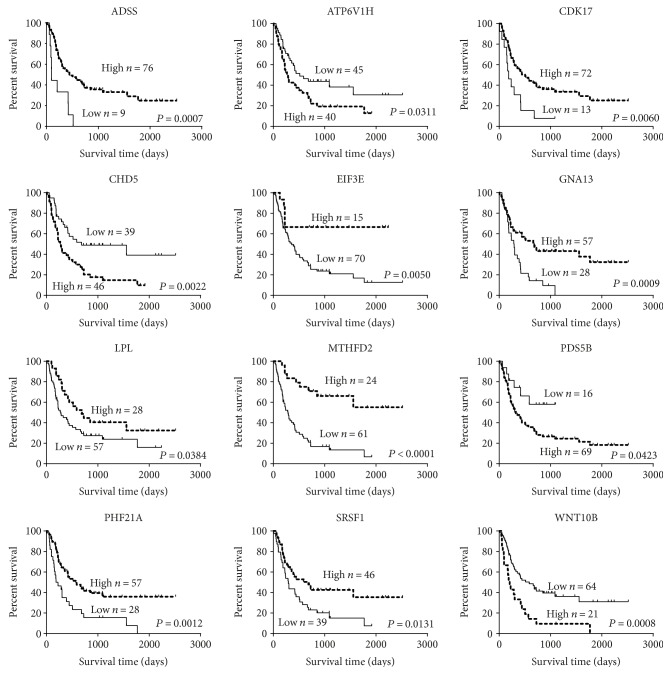
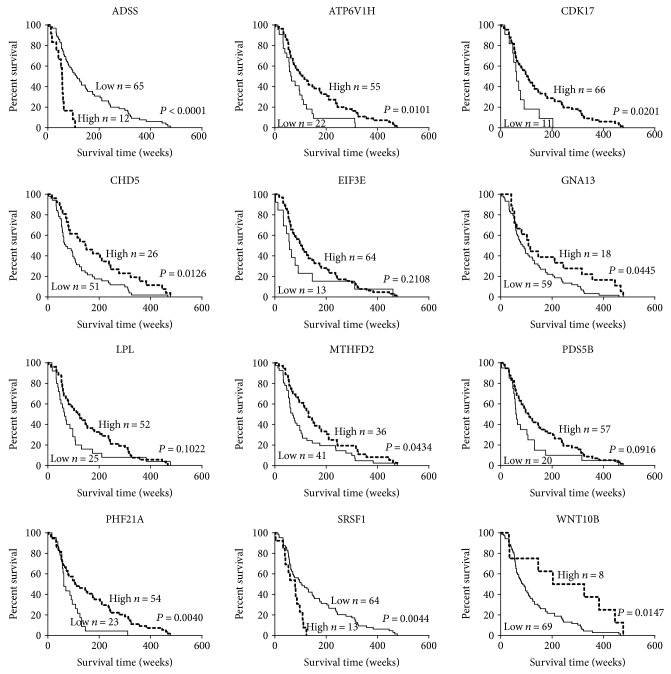
In order to identify biomarkers, we calculated the survival rate for two groups of 12 hub genes in the generation dataset (GSE4412) and compared the result with the validation dataset (GSE4271) through Kaplan-Meier analysis and log-rank test. According to the analysis, we found that only the downregulation of CDK17, GNA13, PHF21A, and MTHFD2 was closely associated with a decreased overall survival among patients with glioma (Figures 3 and 4). The remaining 8 biomarkers had no statistical significance between gene expression and clinical outcome of glioma or no recoverability in the validation dataset. Furthermore, using the Cox proportional hazards model, a multivariate analysis was performed identifying that expression levels of CDK17, PHF21A, and MTHFD2 were independent prognostic factors (Table 4).
Figure 3.
Kaplan-Meier analysis of overall survival for 12 hub genes in the generation dataset of 85 cases.
Figure 4.
Kaplan-Meier analysis of overall survival for 12 hub genes in the validation dataset of 77 cases.
Table 4.
Cox multivariate analyses of biomarkers associated with OS in the generation and validation datasets.
| Dataset | Parameter | Regression coefficient | P value | Risk ratio | 95% confidence interval |
|---|---|---|---|---|---|
| Generation | CDK17 | −0.882 | 0.011 | 0.414 | 0.210 ~ 0.815 |
| MTHFD2 | −1.264 | 0.001 | 0.283 | 0.133 ~ 0.598 | |
| PHF21A | −0.671 | 0.018 | 0.511 | 0.293 ~ 0.891 | |
| Validation | CDK17 | −0.847 | 0.016 | 0.429 | 0.215 ~ 0.856 |
| MTHFD2 | −0.482 | 0.046 | 0.617 | 0.384 ~ 0.992 | |
| PHF21A | −0.620 | 0.024 | 0.538 | 0.314 ~ 0.921 |
4. Discussion
In the present study, we identified DEGs between glioma and normal samples and used a series of bioinformatics analyses to screen key gene and pathways associated with cancer. However, GSE4290 dataset contains only limited number of control samples, 23 out of 180 samples. In order to improve the statistical power of DEG, we defined that the absolute value of the logarithm (base 2) fold change (logFC) greater than 2 and 775 DEGs was obtained. Bioinformatics analysis on DEGs, including GO enrichment, KEGG pathway, PPI network, and survival analyses, found glioma-related genes and pathways that play an important role in cancer initiation and progression.
GO term enrichment analysis demonstrated that 775 downregulated DEGs were significantly enriched in functions involving cellular protein modification, regulation of cell communication, and regulation of signaling. Many studies found that the cellular protein modification, including phosphorylation, ubiquitination, and acetylation, can change the cell biology function, influence disease phenotype, affect glioma cell proliferation, invasion, and apoptosis, and regulate the development of glioma [15–17]. And glioma cells can regulate cell communication, through the information passed to the target cells, interact with the receptor, resulting in specific biological effect such as cell proliferation and cytoskeleton changes, and promote glioma progression and angiogenesis [18]. KEGG pathway analysis indicated that the functions of the downregulated genes were enriched in MAPK signaling, endocytosis, oxytocin signaling, and calcium signaling. Zhang et al. [19] demonstrated that the MAPK signaling pathway induces cell apoptosis in glioma cells and the calcium signaling pathway is involved in quiescence, maintenance, proliferation, and migration in glioma cells [20, 21]. PPI network and module analysis found that the first gene module significantly was enriched in synaptic vesicle cycling. Some results indicate that interference synaptic vesicle cycling could disrupt synaptic function and homeostasis, which would lead to cognitive decline and neurodegeneration in Alzheimer's disease [22]. Therefore, monitoring these signaling pathways may help in the prediction of tumor occurrence and progression.
Since no survival data about GSE4290 could be available, two independent glioma datasets GSE4412 and GSE4271 were applied to detect whether the hub gene could affect the survival time of glioma patients. Survival analysis found that CDK17, GNA13, PHF21A, and MTHFD2 are closely associated with glioma. CDK17 is a member of the cyclin-dependent kinase family. Chaput et al. [23] found that the expression levels of CDK17 are significantly increased in Alzheimer's disease and are associated with the mechanism to promote AD neurodegeneration and may inhibit the pathology development in AD, and Demirkan et al. [24], through a genome-wide association study, found that the CDK17 can be mapped to the glycerophospholipid metabolism pathway. GNA13, one member of the G protein family, is involved in metastasis of tumor cells [25], angiogenesis, and cellular responses to chemokines [26]. In neuronal cells, GNA13 affects neurite outgrowth together with Rho, which is closely related with cell motility and differentiation [27]. Furthermore, GNA13 is coupled to brain-specific angiogenesis inhibitor-1 (BAI1), which is an adhesion-related GPCR, and regulates synaptic function via Rho signaling [28]. PHF21A (also known as BHC80), a plant homeodomain finger-containing protein, can affect the neurofacial and craniofacial development and suppression of the latter and lead to both craniofacial abnormalities and neuronal apoptosis [29]. Moreover, PHF21A specifically binds H3K4me, which is a transcribed genomic locus of regulated posttranslational modification, and implicated the development and maintenance of neural connections [30]. MTHFD2 (methylenetetrahydrofolate dehydrogenase 2) is a mitochondrial enzyme with methylenetetrahydrofolate dehydrogenase and methenyltetrahydrofolate cyclohydrolase activities and has an effect on cancer cell proliferation [31], migration, and invasion [32]. In the expression level of human tumors, MTHFD2 is overexpressed in most cancer types, but exceptions are found in glioma [33], similar to our results. Up to now, the biological functions of CDK17, GNA13, PHF21A, and MTHFD2 in glioma have remained unclear. However, our study shows that the expression level of four key genes are all downregulated in glioma, after comparison with normal brain tissue, and the downregulation is associated with poorer prognosis, as the patients with extended survival time have increased expression of these genes.
At the moment, there are some related bioinformatic research reports of GSE4290 in glioma. Some studies have shown that different enrichment pathway analyses of DEGs can be classified according to their degrees of differential expression during tumor progression in order to explore the deterioration of low into high grade glioma [34]. Some research finds that the DEGs are regulated by transcription factors in glioblastoma [35] and microarray technology has been used to identify the DEGs and their functions in the development of three types of glioma (astrocytoma, glioblastoma, and oligodendroglioma) [36]. Different from these, our study selects the node of the highest score from each module as hub genes in MCODE after comparing nontumor samples with glioma samples. These hub nodes are the key genes of interaction, in the PPI network, that may play important roles in the occurrence and development of glioma. Moreover, hub gene identification is more persuasive, since we validate the association of hub genes and glioma by using survival analysis in two independent datasets to identify four genes that may be cancer biomarkers for glioma. Though not all hub genes associated with the survival of glioma patients, but some hub genes play important roles in immune or inflammation. For example, WNT10B plays an important role in regulating asthmatic airway inflammation through modification of the T cell response [37].
In conclusion, we presumed these key genes identified by a series of bioinformatics analyses on DEGs between tumor samples and normal samples, probably related to the development of glioma. These hub genes could also affect the survival time of glioma patients as validated from two independent datasets. These identified genes and pathways provide a more detailed molecular mechanism for underlying glioma initiation and development. According to the study, downregulation of CDK17, GNA13, PHF21A, and MTHFD2 can be considered as biomarkers or therapeutic targets for glioma. However, further molecular and biological experiments are required to confirm the functions of the key genes in glioma.
Acknowledgments
This work was supported by grants from the National Natural Science Foundation of China (nos. 81672473 and 81502138), the Science and Technology Program of Guangdong (nos. 2014A030310390 and 2017A030313181), the Department of Education of Guangdong Government under the Top-tier University Development Scheme for Research and Control of Infectious Diseases (2016034).
Conflicts of Interest
All authors declare that they have no conflict of interests to state.
Authors' Contributions
Mingfa Liu and Zhennan Xu contributed equally to this work.
References
- 1.Ostrom Q. T., Gittleman H., Liao P., et al. CBTRUS statistical report: primary brain and central nervous system tumors diagnosed in the United States in 2007–2011. Neuro-Oncology. 2014;16:v1–v63. doi: 10.1093/neuonc/nou223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Louis D. N., Perry A., Reifenberger G., et al. The 2016 World Health Organization classification of tumors of the central nervous system: a summary. Acta Neuropathologica. 2016;131(6):803–820. doi: 10.1007/s00401-016-1545-1. [DOI] [PubMed] [Google Scholar]
- 3.Stupp R., Hegi M. E., Mason W. P., et al. Effects of radiotherapy with concomitant and adjuvant temozolomide versus radiotherapy alone on survival in glioblastoma in a randomised phase III study: 5-year analysis of the EORTC-NCIC trial. The Lancet Oncology. 2009;10(5):459–466. doi: 10.1016/S1470-2045(09)70025-7. [DOI] [PubMed] [Google Scholar]
- 4.Freije W. A., Castro-Vargas F. E., Fang Z., et al. Gene expression profiling of gliomas strongly predicts survival. Cancer Research. 2004;64(18):6503–6510. doi: 10.1158/0008-5472.CAN-04-0452. [DOI] [PubMed] [Google Scholar]
- 5.De Preter K., Vermeulen J., Brors B., et al. Accurate outcome prediction in neuroblastoma across independent data sets using a multigene signature. Clinical Cancer Research. 2010;16(5):1532–1541. doi: 10.1158/1078-0432.CCR-09-2607. [DOI] [PubMed] [Google Scholar]
- 6.Kim Y. W., Liu T. J., Koul D., et al. Identification of novel synergistic targets for rational drug combinations with PI3 kinase inhibitors using siRNA synthetic lethality screening against GBM. Neuro-Oncology. 2011;13(4):367–375. doi: 10.1093/neuonc/nor012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cheng W., Ren X., Zhang C., et al. Bioinformatic profiling identifies an immune-related risk signature for glioblastoma. Neurology. 2016;86(24):2226–2234. doi: 10.1212/WNL.0000000000002770. [DOI] [PubMed] [Google Scholar]
- 8.Ward A., Balwierz A., Zhang J. D., et al. Re-expression of microRNA-375 reverses both tamoxifen resistance and accompanying EMT-like properties in breast cancer. Oncogene. 2013;32(9):1173–1182. doi: 10.1038/onc.2012.128. [DOI] [PubMed] [Google Scholar]
- 9.Saura M., Marquez S., Reventun P., et al. Oral administration of bisphenol A induces high blood pressure through angiotensin II/CaMKII-dependent uncoupling of eNOS. The FASEB Journal. 2014;28(11):4719–4728. doi: 10.1096/fj.14-252460. [DOI] [PubMed] [Google Scholar]
- 10.Gene Ontology Consortium. The gene ontology (GO) project in 2006. Nucleic Acids Research. 2006;34(90001) supplement 1:D322–D326. doi: 10.1093/nar/gkj021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kanehisa M., Goto S. KEGG: Kyoto Encyclopedia of Genes and Genomes. Nucleic Acids Research. 2000;28(1):27–30. doi: 10.1093/nar/28.1.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dennis G., Sherman B. T., Hosack D. A., et al. DAVID: database for annotation, visualization, and integrated discovery. Genome Biology. 2003;4(5, article P3) [PubMed] [Google Scholar]
- 13.Szklarczyk D., Franceschini A., Wyder S., et al. STRING v10: protein-protein interaction networks, integrated over the tree of life. Nucleic Acids Research. 2015;43(D1):D447–D452. doi: 10.1093/nar/gku1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Camp R. L., Dolled-Filhart M., Rimm D. L. X-tile: a new bio-informatics tool for biomarker assessment and outcome-based cut-point optimization. Clinical Cancer Research. 2004;10(21):7252–7259. doi: 10.1158/1078-0432.CCR-04-0713. [DOI] [PubMed] [Google Scholar]
- 15.Yoo Y. D., Lee D. H., Cha-Molstad H., et al. Glioma-derived cancer stem cells are hypersensitive to proteasomal inhibition. EMBO Reports. 2017;18(9):p. 1671. doi: 10.15252/embr.201744761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Li X., Yu W., Qian X., et al. Nucleus-translocated ACSS2 promotes gene transcription for lysosomal biogenesis and autophagy. Molecular Cell. 2017;66(5):684–697.e9. doi: 10.1016/j.molcel.2017.04.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fang X., Zhou W., Wu Q., et al. Deubiquitinase USP13 maintains glioblastoma stem cells by antagonizing FBXL14-mediated Myc ubiquitination. The Journal of Experimental Medicine. 2017;214(1):245–267. doi: 10.1084/jem.20151673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kigel B., Rabinowicz N., Varshavsky A., Kessler O., Neufeld G. Plexin-A4 promotes tumor progression and tumor angiogenesis by enhancement of VEGF and bFGF signaling. Blood. 2011;118(15):4285–4296. doi: 10.1182/blood-2011-03-341388. [DOI] [PubMed] [Google Scholar]
- 19.Zhang B., Wu T., Wang Z., et al. p38MAPK activation mediates tumor necrosis factor-α-induced apoptosis in glioma cells. Molecular Medicine Reports. 2015;11(4):3101–3107. doi: 10.3892/mmr.2014.3002. [DOI] [PubMed] [Google Scholar]
- 20.Leclerc C., Haeich J., Aulestia F. J., et al. Calcium signaling orchestrates glioblastoma development: facts and conjunctures. Biochimica et Biophysica Acta (BBA) - Molecular Cell Research. 2016;1863(6):1447–1459. doi: 10.1016/j.bbamcr.2016.01.018. [DOI] [PubMed] [Google Scholar]
- 21.Wypych D., Pomorski P. Calcium signaling in glioma cells – the role of nucleotide receptors. Advances in Experimental Medicine and Biology. 2013;986:61–79. doi: 10.1007/978-94-007-4719-7_4. [DOI] [PubMed] [Google Scholar]
- 22.Lazarevic V., Fieńko S., Andres-Alonso M., et al. Physiological concentrations of amyloid beta regulate recycling of synaptic vesicles via alpha7 acetylcholine receptor and CDK5/calcineurin signaling. Frontiers in Molecular Neuroscience. 2017;10:p. 221. doi: 10.3389/fnmol.2017.00221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chaput D., Kirouac L., Stevens S. M., Jr., Padmanabhan J. Potential role of PCTAIRE-2, PCTAIRE-3 and P-histone H4 in amyloid precursor protein-dependent Alzheimer pathology. Oncotarget. 2016;7(8):8481–8497. doi: 10.18632/oncotarget.7380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Demirkan A., van Duijn C. M., Ugocsai P., et al. Genome-wide association study identifies novel loci associated with circulating phospho- and sphingolipid concentrations. PLoS Genetics. 2012;8, article e1002490 doi: 10.1371/journal.pgen.1002490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rasheed S. A. K., Teo C. R., Beillard E. J., et al. MicroRNA-31 controls G protein alpha-13 (GNA13) expression and cell invasion in breast cancer cells. Molecular Cancer. 2015;14(1):p. 67. doi: 10.1186/s12943-015-0337-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sivaraj K. K., Takefuji M., Schmidt I., Adams R. H., Offermanns S., Wettschureck N. G13 controls angiogenesis through regulation of VEGFR-2 expression. Developmental Cell. 2013;25(4):427–434. doi: 10.1016/j.devcel.2013.04.008. [DOI] [PubMed] [Google Scholar]
- 27.Nurnberg A., Brauer A. U., Wettschureck N., Offermanns S. Antagonistic regulation of neurite morphology through Gq/G11 and G12/G13. The Journal of Biological Chemistry. 2008;283(51):35526–35531. doi: 10.1074/jbc.M804972200. [DOI] [PubMed] [Google Scholar]
- 28.Stephenson J. R., Paavola K. J., Schaefer S. A., Kaur B., van Meir E. G., Hall R. A. Brain-specific angiogenesis inhibitor-1 signaling, regulation, and enrichment in the postsynaptic density. The Journal of Biological Chemistry. 2013;288(31):22248–22256. doi: 10.1074/jbc.M113.489757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kim H. G., Kim H. T., Leach N. T., et al. Translocations disrupting PHF21A in the Potocki-Shaffer-syndrome region are associated with intellectual disability and craniofacial anomalies. American Journal of Human Genetics. 2012;91(1):56–72. doi: 10.1016/j.ajhg.2012.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Vallianatos C. N., Iwase S. Disrupted intricacy of histone H3K4 methylation in neurodevelopmental disorders. Epigenomics. 2015;7(3):503–519. doi: 10.2217/epi.15.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tedeschi P. M., Vazquez A., Kerrigan J. E., Bertino J. R. Mitochondrial methylenetetrahydrofolate dehydrogenase (MTHFD2) overexpression is associated with tumor cell proliferation and is a novel target for drug development. Molecular Cancer Research. 2015;13(10):1361–1366. doi: 10.1158/1541-7786.MCR-15-0117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lehtinen L., Ketola K., Mäkelä R., et al. High-throughput RNAi screening for novel modulators of vimentin expression identifies MTHFD2 as a regulator of breast cancer cell migration and invasion. Oncotarget. 2013;4(1):48–63. doi: 10.18632/oncotarget.756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nilsson R., Jain M., Madhusudhan N., et al. Metabolic enzyme expression highlights a key role for MTHFD2 and the mitochondrial folate pathway in cancer. Nature Communications. 2014;5:p. 3128. doi: 10.1038/ncomms4128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hu G., Wei B., Wang L., et al. Analysis of gene expression profiles associated with glioma progression. Molecular Medicine Reports. 2015;12(2):1884–1890. doi: 10.3892/mmr.2015.3583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wei B., Wang L., Du C., et al. Identification of differentially expressed genes regulated by transcription factors in glioblastomas by bioinformatics analysis. Molecular Medicine Reports. 2015;11(4):2548–2554. doi: 10.3892/mmr.2014.3094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Li G., Pan W., Yang X., Miao J. Gene co-expression network and function modules in three types of glioma. Molecular Medicine Reports. 2015;11(4):3055–3063. doi: 10.3892/mmr.2014.3014. [DOI] [PubMed] [Google Scholar]
- 37.Trischler J., Shiomi T., Turner D. L., et al. Immune modulation of the T cell response in asthma through Wnt10b. American Journal of Respiratory Cell and Molecular Biology. 2016;54(4):584–593. doi: 10.1165/rcmb.2014-0425OC. [DOI] [PMC free article] [PubMed] [Google Scholar]