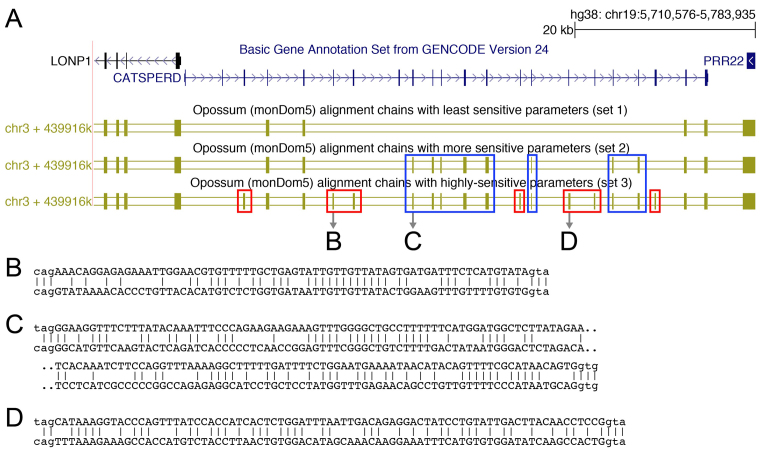
Figure 1.
Alignment parameter sensitivity is crucial to align exons to their orthologous genomic locus. (A) UCSC genome browser screenshot showing the CATSPERD (cation channel sperm associated auxiliary subunit delta) gene locus in the human genome and genome alignments (chains of co-linear local alignments) to opossum computed with three different parameter sets (see text). Several exons of this gene only align to the opossum ortholog with more sensitive parameters (blue boxes) or using a subsequent round of highly sensitive alignments in addition (red boxes). (B–D) Three examples of local alignments covering exons of CATSPERD. Exonic bases are in upper case, intronic bases are in lower case.