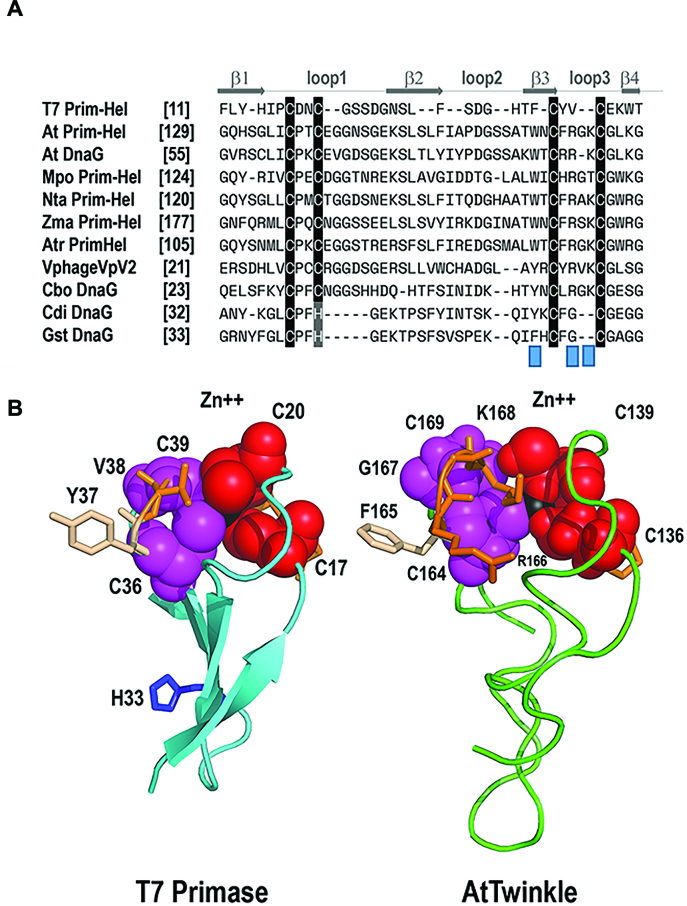
Figure 9.
Structural suggestion for the altered template recognition by AtTwinkle. (A) Multiple sequence alignment between the ZBD of T7 primase-helicase, bacterial primases, and plant Twinkles. The fours cysteines that coordinate the Zn2+ atom in T7 primase can be divided in two CXXC elements. In land plant primases, the first CXXC elements is conserved, whereas the second one is substituted by a CXRXKC element. Protein are designated with abbreviations as follows: Mpo, Marchantia polymorpha; Nta, Nicotiana tabacum; Zma, Zea mays; Atri, Amborella trichopoda; Cbo, Clostridium botulinum; Cdi, Clostridium difficile; Gst, geobacillus stearothermophilus. (B) Computational model of the zinc finger of AtTwinkle in comparison to the crystal structure of T7-Primase. The four β-strands that correspond to the zinc finger are colored in blue. The cysteine residues CXXC element are colored in magenta and red. The side chains of the amino acids in the second CXXC element are in a ball-stick representation. The Zn2+ atom is colored in black. His33 implicated in template recognition in T7 primase is in a ball-stick representation. The computational model of AtTwinkle shows the proposed structural localization of residues R166 and K168.