Figure 4.
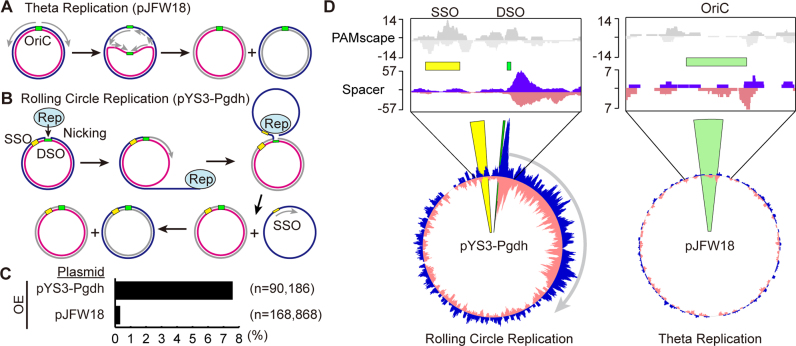
Spacers are preferentially acquired from rolling circle replication plasmids. Both theta (A) and rolling circle (B) replication plasmids were used in these studies. Cartoon models shown here illustrate how replication is thought to occur in each plasmid type. (A) Theta plasmid replication initiates synthesis at a replication origin (OriC). Replication extends the leading strand continuously while synthesizing the lagging strand discontinuously. (B) Rolling circle replication is initiated by a Rep protein, which nicks the double-strand origin (43). Replication proceeds using the un-nicked strand as a template displacing the nicked strand. DNA replication initiates at the single strand origin (SSO) to complete the new double-stranded DNA circle. (C) Spacers from expanded arrays were aligned to both the P. furiosus genome and the plasmid used in the assay. The percentage of spacers aligning to the plasmid rather than the genome (black bar) is indicated for CRISPR7 in of the OE strain. Pooled data from two experiments are presented. (D) Alignment of protospacers to the plasmid shows an unequal distribution. Top panels show 5′-CCN-3′/5′-NGG-3′ PAM scape on each plasmid. Green bars show origin of DNA replication. Bottom panels show the distribution of protospacers across plasmids pYS3-Pgdh and pJFW18. Protospacers on the plus and minus strand are indicated in blue and pink, respectively. Protospacers from CRISPR7 in the OE strain are shown here; other CRISPR arrays and WT strains show similar distributions.