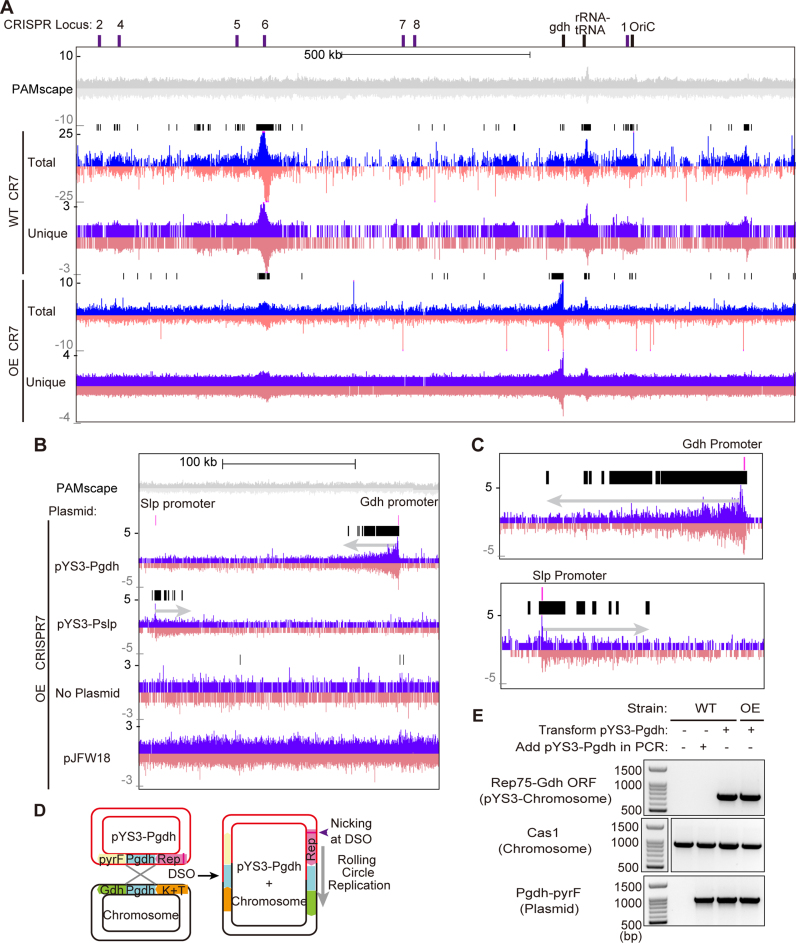
Figure 5.
Aligned protospacers are not evenly distributed across the P. furiosus chromosome. (A and B) Distribution of protospacers on the P. furiosus chromosome. Protospacers on the plus and minus strand are indicated in blue and pink respectively. Protospacers were deduced from aligning new spacers acquired into CRISPR7 loci of WT and OE strains. The 5′-CCN-3′/5′-NGG-3′ PAMscape across the chromosome is shown in gray (top). Black bars show regions where protospacers are significantly enriched over the background, as detected by the peak-calling software from the HOMER package. (A) Distribution of protospacers across the full length of the chromosome. (B and C) Protospacers are enriched around areas where homology exists between the chromosome and the plasmid. Prominent clusters of protospacers are observed extending 1000s of bps out from the region of homology. Protospacers from CRISPR7 in the OE strain are shown here; other CRISPR arrays and WT strains show similar distributions. Close-up panels (C) show that the peak of enrichment starts ∼300bp from the location of homology between the plasmid and chromosome, and extends out from that point. (D) Model of homologous recombination occurring at the homologous region (Pgdh) between plasmids and the P. furiosus chromosome. Recombination results in integration of the plasmid into the chromosome. (E) PCR results confirm that homologous recombination between the plasmid and chromosome occurs. Top panel shows amplified products of plasmid-integrated chromosome. Middle panel shows internal control of chromosome and bottom panel shows internal control of the plasmid. Sizes of DNA standards are indicated.