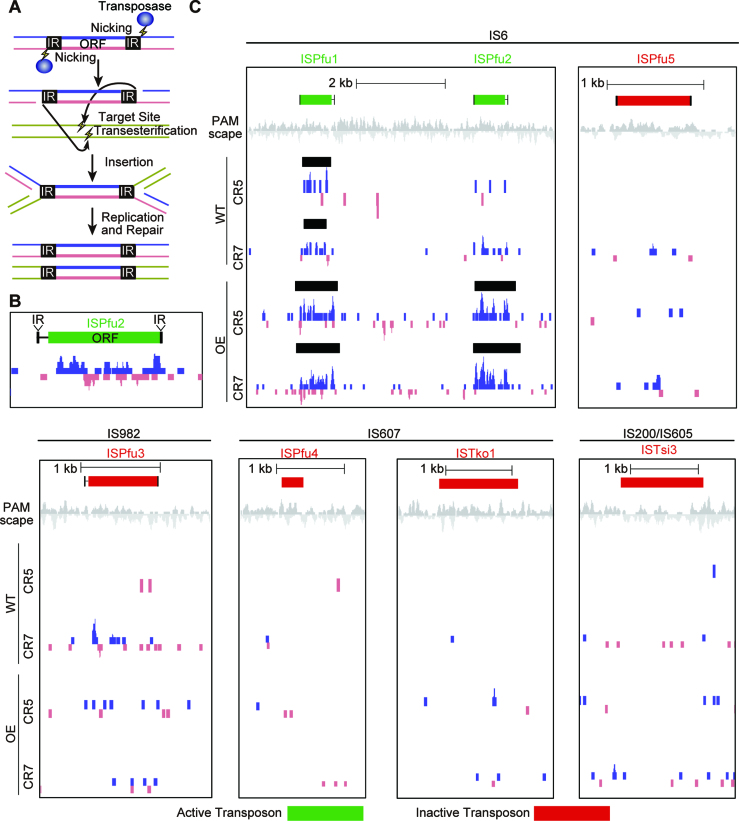
Figure 6.
Protospacers are enriched for transposons. (A) Cartoon model of the mechanism for transposon replication. Transposases nick each transposon at the 3′ end. Target DNA is also nicked and the transposon joins to the target. The transposon intermediate is resolved by recombination. The original and target sites are separated, each harboring one copy of the transposon. (B and C) Protospacers are significantly enriched around active transposons. Protospacers on the plus and minus strand are indicated in blue and pink respectively. Protospacers were deduced by aligning new spacers acquired into the CRISPR5 and seven loci of WT and OE strains. Black bars show areas were protospacers are significantly enriched, as detected by peak finding software in the HOMER package. Transposons that were previously described as active (44) were 10 times more likely to have an associated peak than those described as inactive (red).