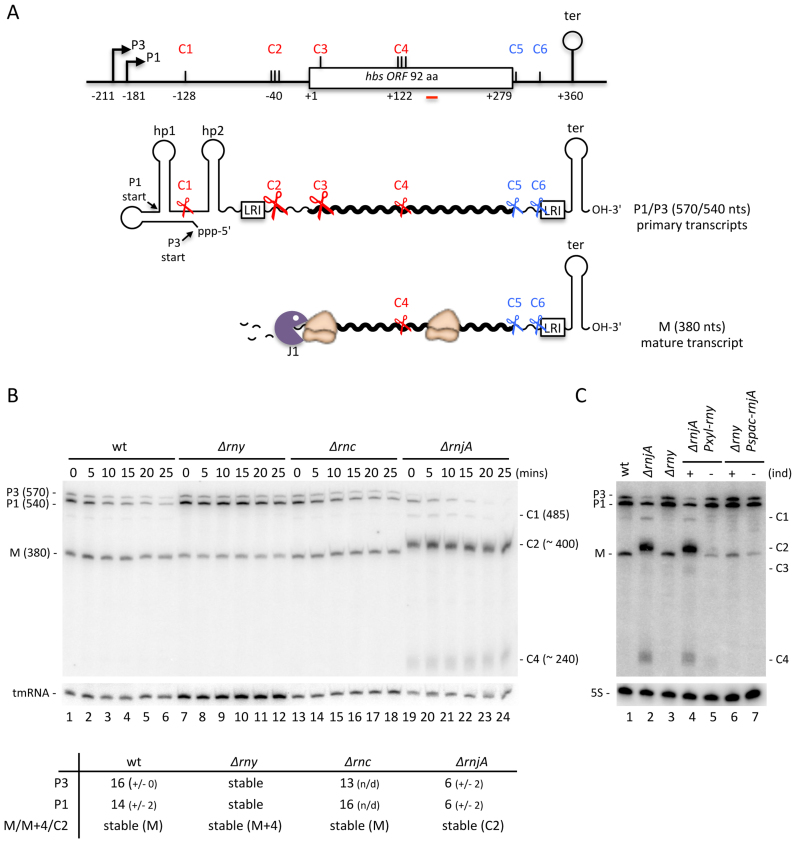
Figure 1.
Endonucleolytic cleavage of the 5′-UTR by RNase Y provides access to the 5′-3′ exoribonuclease RNase J1 for hbs mRNA maturation. (A) Schematic of the B. subtilis hbs gene and transcripts. The P3 and P1 promoters and transcription terminator (ter) are indicated. C1–C6 refer to endonucleolytic cleavage sites mapped in the course of this and previous studies. Complementary sequences involved in a proposed long-range interaction (LRI) between the 5′ and 3′-UTRs are indicated. Transcripts are shown as wavy lines. Hp1 and hp2 refer to 5′ hairpin structures. Key endonucleolytic cleavage events are symbolized by scissors; the 5′-3′ exonuclease RNase J1 by a Pacman symbol. The positions of different features are given relative to the translation start site, designated as +1. (B) Northern blot of total RNA isolated from wild-type (wt), RNase Y (Δrny), RNase III (Δrnc) and RNase J1 (ΔrnjA) mutants at different times after rifampicin addition. Calculated half-lives of the major transcripts are given underneath the blot. Half-lives >30 min are referred to as stable. Average decay curves are shown in Supplementary Figure S1. (C) Northern blot of total RNA isolated from wt, ΔrnjA strains, Δrny and ΔrnjA Pxyl-rny and Δrny Pspac-rnjA depletion strains grown in the presence (+) or absence (–) of inducer (ind). For panels B and C, the migration positions of the P3 and P1 primary transcripts, the mature species (M) and the degradation intermediates cleaved at C1, C2, C3 and C4 are indicated. Blots were first probed with oligo CC287 (hbs), shown as a red line in panel A, and then reprobed with oligos CC1444 (tmRNA) or HP246 (5S rRNA) as a loading control. Number of biological replicates (n) = 2.