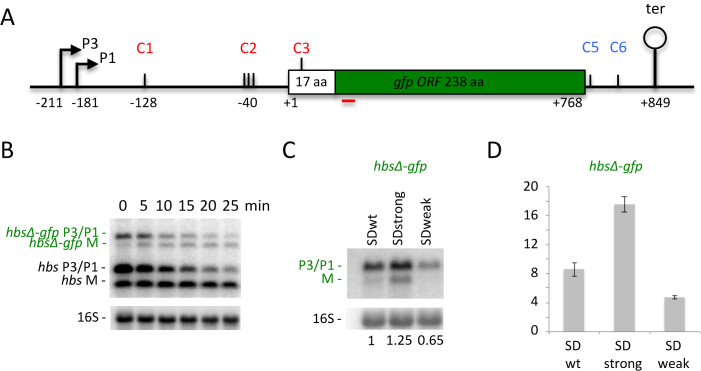
Figure 4.
Ribosomes can efficiently escape the exceptionally strong SD sequence of the hbs mRNA. (A) Schematic of the B. subtilis hbsΔ-gfp construct. Labeling is as in Figure 1. (B) Northern blot of total RNA isolated from strains carrying the hbsΔ-gfp WT construct integrated into the chromosome in amyE at different times after rifampicin addition (n = 2). The blot was probed with oligos CC1708 (gfp; shown as a red line in panel A), CC287 (to reveal the native hbs mRNA) and then CC058 (16S rRNA) as a loading control. (C) Northern blot of total RNA isolated from strains carrying the hbsΔ-gfp SDwt, hbsΔ SDstrong and hbsΔ SDweak constructs (n = 2). The blot was probed with CC1708 (gfp) and then CC058 (16S rRNA) as a loading control. Ratios of total hbsΔ-gfp transcripts (P3+P1+M) to 16S rRNA are indicated under the blot. (D) GFP expression levels normalized to OD600 for the strains containing the hbsΔ-gfp SDwt, hbsΔ SDstrong and hbsΔ SDweak constructs (n = 3). Measurements were taken at OD600 = 1.