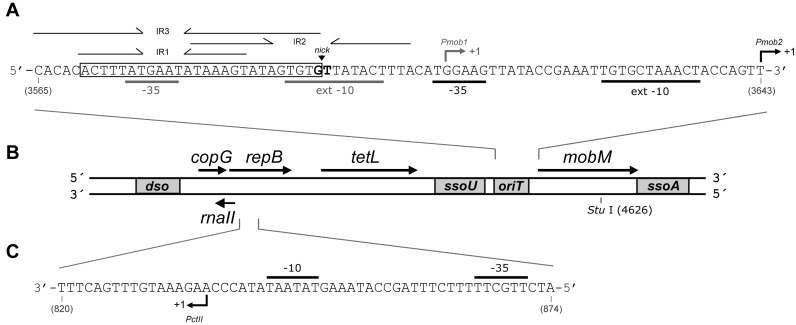
Figure 1.
Genetic map of plasmid pMV158. Only relevant features are indicated. (A) Nucleotide sequence (coding strand) of the region that includes the origin of transfer (oriT), the promoters of the mobM gene (Pmob1 and Pmob2) and the corresponding transcription initiation sites (arrows). The three inverted repeats (IR1, IR2, IR3), the minimal oriT sequence (coordinates 3570–3595) and the nicking site (nick) are indicated, as well as the −10 and −35 boxes of the Pmob1 and Pmob2 promoters used in Lactococcus lactis (72) and in Escherichia coli (45), respectively. (B) Genes encoded by the three pMV158 modules: (i) copG, repB and rnaII, involved in replication and its control; (ii) tetL, a type L tetracycline-resistance determinant and (iii) mobM, involved in mobilization. The origins for leading-strand (dso) and lagging-strand (ssoU, ssoA) synthesis are boxed, as well as the oriT. The position of the single StuI restriction site (coordinate 4626) is indicated. (C) Nucleotide sequence (non-coding strand) of the region (coordinates 820–874) that includes the promoter of the rnaII gene (PctII). The −10 and −35 sequences, and the transcription start site (arrow) (46) are indicated.