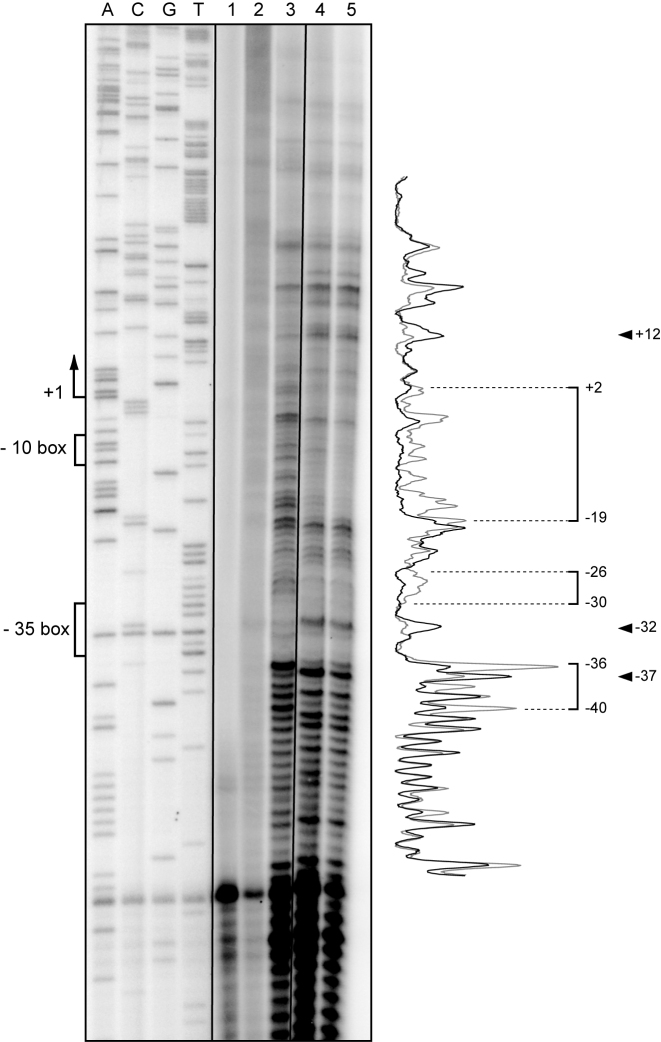
Figure 3.
DNase I footprinting analysis on supercoiled pAS DNA. Plasmid pAS was incubated with MobM. Binding reactions were treated with DNase I and then subjected to primer extension using the 32P-labelled FP-rnaII oligonucleotide. The primer extension reactions were performed on plasmid pAS in the absence of both MobM and DNase I (lane 1), in the presence of MobM (2 μM) and absence of DNase I (lane 2), in the absence of MobM and presence of DNase I (lane 3), and in the presence of both MobM (1 and 2 μM) and DNase I (lanes 4 and 5, respectively). Dideoxy-mediated chain-termination sequencing reactions using pAS DNA and the FP-rnaII oligonucleotide were run in the same gel (lanes A, C, G and T). The −10 and −35 boxes of the PctII promoter and the MobM-protected regions are indicated with brackets. The indicated positions are relative to the transcription start site (position +1) of the PctII promoter. Arrowheads indicate positions that are more sensitive to DNase I cleavage. Densitometer scans corresponding to DNA without MobM (lane 3, grey line) and DNA with MobM (lane 5, black line) are shown.