Abstract
Two Telomeric Associated Sequences, TAS-R and TAS-L, form the principal subtelomeric repeat families identified in Drosophila melanogaster. They are PIWI-interacting RNA (piRNA) clusters involved in repression of Transposable Elements. In this study, we revisited TAS structural and functional dynamics in D. melanogaster and in related species. In silico analysis revealed that TAS-R family members are composed of previously uncharacterized domains. This analysis also showed that TAS-L repeats are composed of arrays of a region we have named “TAS-L like” (TLL) identified specifically in one TAS-R family member, X-TAS. TLL were also present in other species of the melanogaster subgroup. Therefore, it is possible that TLL represents an ancestral subtelomeric piRNA core-cluster. Furthermore, all D. melanogaster genomes tested possessed at least one TAS-R locus, whereas TAS-L can be absent. A screen of 110 D. melanogaster lines showed that X-TAS is always present in flies living in the wild, but often absent in long-term laboratory stocks and that natural populations frequently lost their X-TAS within 2 years upon lab conditioning. Therefore, the unexpected structural and temporal dynamics of subtelomeric piRNA clusters demonstrated here suggests that genome organization is subjected to distinct selective pressures in the wild and upon domestication in the laboratory.
Keywords: subtelomeres, TAS, Drosophila, piRNA, transposable element
1. Introduction
In the Drosophila melanogaster genome, Telomeric Associated Sequences (TAS) are moderately repeated sequences that correspond to non-coding heterochromatin. They are positioned between euchromatin and the distal telomeric transposon arrays, consisting of non-LTR retrotransposons: HeT-A, TAHRE and TART (HTT), not present elsewhere in the genome.1–4 Due to their position, TAS constitute a transition zone between telomeres and genes.
The initial molecular analysis of TAS repeats comes from the study of a 10-kb fragment isolated from the Dp1187 X minichromosome.1 The telomeric region of this chromosome contains four tandem TAS repeats of 1.8 kb for the longest (X-TAS) and two repeats of 0.9 kb1 (Fig. 1A). Each TAS repeat contains three tandem subrepeats of 172 bp that were later identified as solo LTRs of the Invader4 retrotransposon (INV-4).5
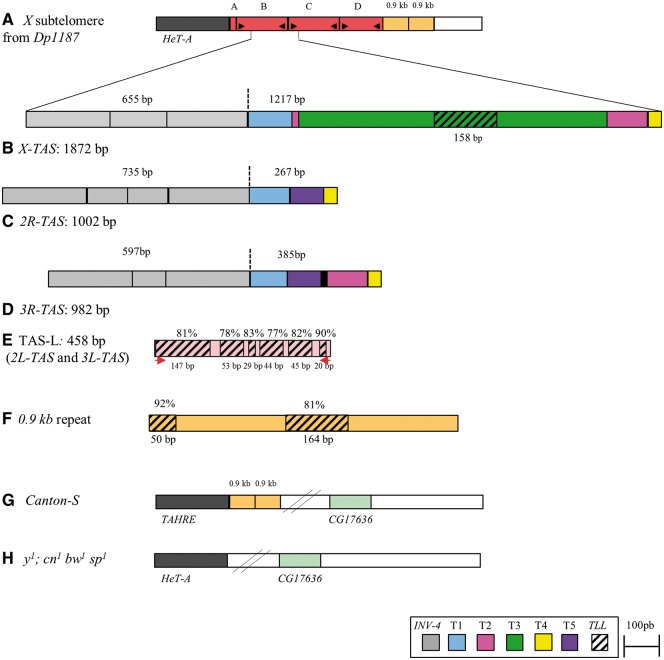
Figure 1.
Subtelomeric regions in Drosophila melanogaster. (A) X subtelomeric region from the Dp1187 minichromosome contains four TAS repeats [TAS-A (205 bp), TAS-B (1,872 bp), TAS-C (1,858 bp) and TAS-D (1,575 bp)] located between telomeric elements (HeT-A) and two 0.9 kb repeats. (B–D) Schematic representation of the TAS-R family repeats represented by TAS-B for X-TAS (B), 2R-TAS (C) and 3R-TAS (D) located on the X, second and third chromosomes, respectively. They are composed of several domains [INV-4 solo LTR, T1, T2, T3, T4 and T5 domain]. In order to simplify the comparison between members of TAS-R family, the starting point for each TAS repeat was arbitrary positioned at the beginning of the INV-4 sequence. (E) TAS-L repeats on 2L and 3L chromosomes are composed of six fragments of a sequence found in the T3 domain of X-TAS repeats, that we named TLL (hatched boxes). (F) The TLL domain is also found in the 0.9-kb repeats adjacent to X-TAS. The T3-TLL sequence (B) is taken as the reference here for determining percentage of identity between sequences (E, F). Identity between the TLL of 164 bp in the 0.9 kb repeat and the TLL of 147 bp in TAS-L is 85%. (G) The X chromosome of the Canton-S strain is lacking X-TAS repeats. (H) The X chromosome of the y1; cn1bw1sp1 line lacks both X-TAS repeats and 0.9 kb repeats. In all schematic representations of telomeric sequences the telomeres are to the left. TAS-R probes used in in situ hybridization were PCR amplified using primers indicated by black arrows (A). Primers used to PCR amplify TAS-L repeats are indicated by arrows (E).
Early in situ hybridization studies on polytene chromosomes and more recent sequence alignments identified common subtelomeric regions, corresponding to the INV-4 LTRs, for the X, 2R and 3R chromosomes (X-TAS, 2R-TAS and 3R-TAS).1,6,7 These three subtelomeric regions have been grouped together as the TAS-R family to differentiate them from the TAS-L family, which is devoid of INV-4 sequences and localized at 2L and 3L telomeres.8 TAS-L are made-up of non-coding repeats.3 Recent publications mentioned sequences identities between the TAS-R and TAS-L families.9,10
X-TAS, a TAS-R family member, has been shown to play a major role in the establishment of P element repression following invasion of the D. melanogaster genome between the 1950s and the 1970s.11–13 The study of this repression led to the discovery of an ovarian silencing mechanism involving PIWI interacting RNA (piRNA) biogenesis called the Trans-Silencing Effect (TSE).14–16 piRNAs are a class of non coding small RNAs of 23–29 nt produced from specific loci, called piRNA clusters.17 piRNA clusters are heterochromatic, mainly consisting of fragments of Transposable Elements (TEs), and involved in repression of TE transposition at transcriptional and/or posttranscriptional levels in animal gonads.18 TAS-R, especially X-TAS, appear to be piRNA clusters, into which P elements (as well as P-derived transgenes) can insert.1,10,19 Once inserted, P sequences are included in the piRNA produced and these piRNA will participate to repression of euchromatic P copies in the germline leading to the so-called TSE.20–23
Current literature studying repression of TEs is dealing with the fact that each TE might be potentially regulated by multiple piRNA clusters at the same time. Here, we focus on a specific class of piRNA clusters, the subtelomeric clusters. We extended the analysis of their structure, function and dynamics in long-term laboratory stocks and flies recently collected from the wild. We further addressed their evolutionary origin by comparative analysis of Drosophilidae genomes. Using in situ hybridization on polytenes chromosomes, PCR amplification, ovarian small RNA sequencing and in silico sequence comparison, we identified a core sequence, that we called TLL, for TAS-L like, in D. melanogaster subtelomeric piRNA clusters. In addition, the TLL sequence was shown to be a conserved part of piRNA clusters in subtelomeric regions of related melanogaster subgroup species. Finally, a striking difference in the presence and absence of X-TAS between long-term laboratory stocks and wild D. melanogaster populations was observed. An extensive analysis of lines established from flies from the wild showed that they all possessed X-TAS, while this was not the case for many laboratory stocks. In addition, a survey over a 2-year period of the wild-type lines allowed us to witness the loss of X-TAS during Drosophila domestication under laboratory conditions. This study reveals an unexpected feature for piRNA clusters that, although fulfilling an essential role as guardians of the genome by repressing TEs, can be lost under artificial laboratory conditions without significantly impairing genome integrity of their hosts.
2. Materials and methods
2.1. Drosophila stocks
The following Fly stocks from the Bloomington stock center were used: y1; cn1bw1sp1 (FBst0002057), Canton-S (FBst0000001), Oregon-R-C (FBst0000005), VAG_2 (FBst0003876), Wild_11C (FBst0003894), BER_2 (FBst0003840) and Wild_5A (FBst0003885). The w1118, P-1152, NA-P(1A) and Lk-P(1A) belong to our laboratory stock collection. P-1152 carries two P-lacZ fusion enhancer trap transgenes (FBti0005700), inserted in X-TAS, that contain an in-frame translational fusion of the E. coli lacZ gene to the second exon of the P transposase gene, as well as the rosy+ gene as a transformation marker.19,24 The Lk-P(1A) line carries two full-length P elements inserted in X-TAS. NA-P(1A) carries a P element inserted in X-TAS that has undergone a telomeric deletion leaving a 5′ deleted P element. These telomeric insertions have been genetically isolated from inbred lines derived from wild populations collected in Lerik, Azerbaijan, for Lk-P(1A)13 and from Nasr’Allah, Tunisia, for NA-P(1A)25 in a genome devoid of other P elements. The Drosophila sechellia strain was kindly provided by Aurélie Hua-Van (EGCE, CNRS-IRD-University Paris-Sud-University Paris Saclay). Between 2013 and 2015, we collected Drosophila populations from the wild in Europe (France and Austria), Africa (Ethiopia and Senegal) and South America (Brazil and French Guiana). In French Guiana, flies were collected in Saul, an isolated town in the middle of the rainforest. The strains collected in 2013 in Marsais (France) and in 2012 in Gotheron (Valence, France) were kindly provided by Jean-Michel Gibert (LBD, IBPS, UPMC) and Cristina Vieira (LBBE, CNRS, University Lyon 1), respectively.
2.2. Sequence accession numbers
The following sequences were used in this study: X-TAS (L03284),1 2L-TAS (U35404),3Canton-S cosmid (AL031884),26 clone AF520587,27Invader 4 (FBgn0063427), 2R-TAS and 3R-TAS recovered from Lin and Yin, 20077 and y1; cn1bw1sp1 from FlyBase.org.28
2.3. Preparation of polytene chromosomes and fluorescence in situ hybridization
Salivary glands from four to eight third instar larvae grown at 18°C were dissected and fixed in 45% acetic acid. After squashing, slides were quick-frozen in liquid nitrogen and dehydrated in ethanol (90%). Slides were then washed 30 min in 2× SSC at 65°C and dehydrated in ethanol baths (90%). Before hybridization, chromosomal DNA was denatured in NaOH 0.07N. Probes were prepared from PCR amplification products using primers allowing amplification of full length TAS-B repeat of 1.8 kb from the P-1152 genome (Fig. 1), a full length 2.9–kb P element from the Lk-P(1A) genome and the 855 bp repeats from the D. sechellia genome for in situ hybridization experiments in Fig. 2. Except for the 855 bp repeats, these PCR products were then cloned into the pCR2.1 plasmid (Thermofisher Scientific) for preparing either biotinylated DNA probes using the BioNick Labelling System (Invitrogen) and digoxigenin probes using the DIG-Nick Translation Mix (Roche). Slides were incubated with 1 μg of probe at 37°C overnight. Slides were washed two times in 2× SSC at 37°C. For detection, we used FITC-extravidine at 50 μg/ml (Sigma) and anti-DIG-rhodamine at 4 μg/ml (Roche) final concentrations. Slides were stained in DAPI-VectaShield mounting medium and observed using a Leica TCS SP5 confocal microscope system.
Figure 2.
Subtelomeric piRNA clusters in the melanogaster subgroup. (A) D. sechellia Scaffold 31 contains five complete 855 bp repeats. Each repeat is composed of two subrepeats (191 and 138 bp) with sequence identities to D. melanogaster TLLs (hatched boxes), one 129 bp sequence derived from TLL (hatched boxes) (Supplementary Fig. S7) and two unrelated sequences of 274 bp and 123 bp. (B) D. simulans Scaffold 3R contains a 403-bp region composed of one TLL and two sequences of 105 and 129 bp displaying sequence identity with the 129 bp of D. sechellia followed by a T2 and a T5 domains. (C) In situ hybridizations on D. sechellia, D. simulans (Chantemesle) and D. melanogaster (Oregon-R-C) polytene chromosomes were performed using the D. sechellia 855 bp repeats as a probe. The probe was PCR amplified using primers shown by arrowheads in (A). Probe hybridizations at telomeres indicated by arrowheads are consistent with the in silico analysis. Labelling at the D. melanogaster centromeres can also be detected corresponding to smaller domains of identity (see text). (D–E) TLL-containing domains identified in D. sechellia and D. simulans are part of subtelomeric piRNA clusters. Normalized germline small RNAs were mapped to each reference sequences: the 855 bp D. sechellia repeat, the 403 bp D. simulans sequence and the 158 bp-TLL D. melanogaster sequences. (D) Size distribution histograms indicate that the majority of small RNAs produced from telomeric regions in ovaries of the three Drosophila species are between 23 and 29 nt in length. The number in each panel is the proportion of 23–29 nt beginning with a 5′ uridine (1 U bias). A bias exists toward sense (plus strand) vs. antisense (minus strand) strand production. (E) Readmaps show the abundance and the distribution of the piRNAs mapping to each of the reference sequences.
2.4. Sequence analysis
Sequence comparisons were performed using BLAST (NCBI) and SeaView using MUSCLE for multiple sequence alignments.29 Minor manual gap adjustments were introduced when needed.
2.5. DNA extraction and PCR amplification
Flies were manually homogenized in a buffer containing 0.1-M Tris pH9, 0.1-M EDTA pH8 and 1% SDS and then incubated at 70°C for 30 min. Potassium Acetate 8 M and isopropanol were added prior centrifugation. Pellets of DNA were washed and resuspended in water and PCR was performed using primers (available upon request). This protocol was used for DNA extraction from single flies (Tables 2 and 3, Supplementary Tables S2 and S3) as well as from groups of twenty to fifty flies (Fig. 3 and Supplementary Table S2). Since the absence of a PCR amplification product could be attributed to poor DNA quality, we confirmed the amplificability of DNA samples using independent sets of primers (data not shown).
Table 2.
Distribution of TAS family in recently collected strains
| Year of capture | T3 domaina | X-TASb | 2R-TASb | 3R-TASb | TAS-La | |
|---|---|---|---|---|---|---|
| Europe | ||||||
| Paris | 2013 | + | + | + | − | + |
| Combs-la-Ville | 2013 | + | + | + | + | + |
| Viry-Châtillon-3 | 2013 | + | + | + | + | + |
| Chantemesle-iso | 2013 | + | + | + | + | + |
| Marsais-53Ja | 2013 | + | + | + | + | + |
| Marsais-80J | 2013 | + | + | + | − | + |
| Marsais-86C | 2013 | + | + | + | + | + |
| Marsais-87C | 2013 | + | + | + | − | + |
| Niort-A1 | 2013 | + | + | − | + | + |
| Niort-G3 | 2013 | + | + | + | − | + |
| Niort-P1.3 | 2013 | + | + | − | + | + |
| Niort-P2.1 | 2013 | + | + | − | + | + |
| Reth | 2013 | + | + | + | + | + |
| Mauzé Thouarsais-2 | 2013 | + | + | − | + | + |
| Mauzé Thouarsais-3 | 2013 | + | + | − | + | + |
| Sanguinet-1.5 | 2014 | + | + | + | + | + |
| Rouans1 | 2013 | + | + | + | + | + |
| Gotheron, Valence-iso3 | 2012 | + | + | + | − | + |
| Africa | ||||||
| Addis Ababa 2 | 2014 | + | + | + | + | + |
| Kafountine | 2015 | + | + | + | − | + |
| South America | ||||||
| Saul, Cacao H2A2 | 2014 | + | + | + | − | + |
| Saul, Cacao H1C | 2014 | + | + | + | + | + |
| Ilha Grande 1, Rio Janeiro | 2014 | − | − | + | + | + |
aThe data were obtained by PCR amplification using specific primers testing the presence of T3 domain or TAS-L family.
bThe data were obtained by in situ hybridization on polytene chromosome of at least four larvae using a TAS-R probe.
Table 3.
X-TAS loss in Drosophila stocks within their first two years in laboratory
| Origin | Year | Few generations after trapping from wild | Stock A |
Stock B |
||
|---|---|---|---|---|---|---|
| Tested 1 year after collection | Tested 2 years after collection | Tested 1 year after collection | Tested 2 years after collection | |||
| France | ||||||
| Marsais-53Ja | 2013 | ± | + | + | + | − |
| Marsais-90C | 2013 | NT | + | + | + | − |
| Niort-G2 | 2013 | NT | + | − | + | + |
| Niort-P2.4 | 2013 | NT | + | − | + | + |
| Niort-P2.6 | 2013 | NT | + | NT | + | − |
| Niort-L3 | 2013 | NT | NT | NT | + | − |
| French Guiana | ||||||
| Saul FG24 | 2013 | + | + | NT | + | − |
| Saul A3 | 2014 | + | − | NT | + | NT |
| Saul 13 | 2014 | + | + | NT | − | NT |
“+” when PCR amplifications were obtained, "-" when no PCR amplifications were obtained.
Please refer to Supplementary Table S2 for a complete view of results for all the stocks tested.
NT: Not Tested.
Figure 3.
Schematic representation of X subtelomeric regions after X-TAS loss. Four types of X subtelomeres were identified: (A) with one each of the TAS-B, -C, -D and the 0.9 kb repeats like the Dp1187 minichromosome1; (B) with only theTAS-D repeat and the 0.9 kb repeats like NA-P(1A)25; (C) with only the 0.9 kb repeats like Canton-S; (D) with neither TAS repeats nor the 0.9 kb repeats like y1; cn1bw1sp1. Stocks found to be devoid of X-TAS repeats by in situ hybridization and/or PCR amplification were tested to identify their X subtelomeric structure. This was assayed by PCR amplifications using specific primers for TAS-B (see arrows under), TAS-D (see arrows) and the 0.9 kb repeats (see arrows) (A). Noticeably, the 0.9 kb repeats were systematically amplified in stocks that possess TAS-B and/or TAS-D. The 0.9 kb repeats can also be stably maintained on their own at the tip of the X chromosome as observed for the Canton-S strain. Strains shown to have recently lost X-TAS (see Table 3) are marked by an asterisk (*). Loss of X-TAS might occur by telomeric terminal breakage rather than by internal deletion that might be repaired by transposition of new telomeric HTT (HeT-A, TAHRE).
2.6. Small RNA sequencing
Total RNA extraction, small RNA library preparation and sequencing were performed as described in Hermant et al.30 Sequence reads in FASTQ format were trimmed from the adapter sequence 5′-TGGAATTCTCGGGTGCCAAG-3′ and matched to reference sequences (TAS, P element, or D. sechellia repeats) using Bowtie.31 Multiple mappers from 23 to 29 nt matching the reference sequences with zero mismatches were retained for subsequent analysis. Annotation of small RNA libraries and normalization factors are described in Supplementary Table S1. Sequence length distributions and small RNA mapping were generated from Bowtie alignments using Python and R (www.r-project.org/) scripts, which were wrapped and run in a Galaxy instance publicly available at http://plastissipi.fr. Tools and workflows used in this study can be downloaded from this Galaxy instance.
2.7. Data availability
Small RNA sequences have been deposited at the European Nucleotide Archive (ENA) under accession number PRJEB19350.
3. Results and conclusions
3.1. Structural analysis of TAS-R family members reveals that they are more highly related than previously suspected
X, 2R and 3R chromosomal arms in D. melanogaster contain subtelomeric TAS-R sequences called X-TAS, 2R-TAS and 3R-TAS, respectively, though not all strains present all three. Despite their fundamental role as piRNA clusters involved in repression of TEs (such as P element), little is known about their dynamics. To address this question, we first revisited the precise structure of these TAS.
The X subtelomeric region was studied by revisiting the Dp1187 X minichromosome sequence encoding the only complete X-TAS locus sequenced to date.1 According to this initial study, it contains a tandem of four TAS repeats, named TAS-A to -D, upstream of two 0.9 kb repeats (Fig. 1A). TAS-B (1,872 bp), the longest, was considered to be the reference for X-TAS repeats.1TAS-A (205 bp) appears to originate from a telomeric 5′ terminal deletion within a longer TAS repeat repaired by telomeric HTT transposition events. TAS-C (1,858 bp) differs by a few indels from TAS-B. TAS-D (1,575 bp, 95% identical with TAS-B) contains a 313-bp deletion within the 3′ region of the unit compared to TAS-B (Supplementary Fig. S1). To revisit the portion of TAS-R originating from INV-4 LTRs, we performed sequence comparisons between the LTR of the canonical INV-4 346 bp sequence and TAS-R sequences. We found that the portion of the TAS-R that matches with the INV-4 LTR were more extensive than the three 172 bp repeats originally described.1,5 For instance, the three partial INV-4 LTR sequences identified in X-TAS were 248, 172 and 235 bp in length with more than 83% similarity with the canonical LTR. 2R-TAS was shown to contain four tandem INV-4 LTR partial sequences and 3R-TAS, three INV-4 LTR partial sequences, two of which were longer than 172 bp (for details see Fig. 1B–D and Supplementary Fig. S2).
We next asked whether the shared origin between the X-TAS, 2R-TAS and 3R-TAS was limited to the INV-4 LTR region. By performing multiple alignments between the remnants of the three loci, five other domains were identified that we named T1, T2, T3, T4 and T5 (Fig. 1B–D). The T1 and T4 domains were present in all three TAS-R loci, the T2 domain in X-TAS and 3R-TAS, and the T5 domain in 2R-TAS and 3R-TAS (Supplementary Fig. S3). Only the T3 domain, 909 bp in length, was found to be restricted to one TAS-R family member, X-TAS, except for an internal domain (see below).
Taken together, the characterization of TAS-R sequences indicates that members of this family are more closely related to each other than originally suspected. Indeed, they are a mosaic of six domains (T1–T5 and INV-4) organized in a specific order at subtelomeres.
3.2. TAS-R and TAS-L families are related
We next investigated the origin of each of the “T”domains (T1–T5) identified above. Using several databases, the only sequence alignments recovered corresponded to those of a 158-bp region located in the T3 domain with sequences in subtelomeric TAS-L family repeats located on the 2L and 3L chromosomes (Fig. 1B and E, Supplementary Fig. S4) as well with sequences in the two domains of the 0.9 kb repeats next to X-TAS (Fig. 1B and F and Supplementary Fig. S5). The 2L-TAS and 3L-TAS are identical subtelomeric minisatellite repeats.3 In the y1; cn1bw1sp1 reference genome (FlyBase.org), we found 11 repeats and 42 repeats of 458 bp on the 2L and 3L subtelomeres, respectively (Supplementary Fig. S6). Such similarities between TAS-R and TAS-L families have been suggested recently.9,10 In this study, we specify these similarities showing that TAS-L repeats consists of six tandemly-repeated sequences of 147, 53, 29, 44, 45 and 20 bp in length that share over 80% sequence similarity with the 158 bp region identified in the T3 domain of X-TAS (Fig. 1E). For this reason, we named this 158 bp domain, the TLL domain. BLAST searches using the whole T3 domain also unveiled similarities between TLL and an unmapped clone composed of fragments of TEs isolated from a D. melanogaster strain collected in Samarkand, Uzbekistan (73% identity)27 as well as with two domains of 146 bp in pericentromeric regions of y1; cn1bw1sp1 (∼80% identity, data not shown).
With the identification of the TLL domain within chromosomic regions carrying remnants of TEs, it is tempting to speculate that TLL could be of TE origin. Since the TLL domain was found to be part of a TAS-R member, X-TAS, of the adjacent 0.9 kb repeats and of the TAS-L family repeats, it can be proposed to represent the common ancestor sequence of the main subtelomeres in the D. melanogaster genome.
3.3. TAS sequences in the melanogaster subgroup are likely piRNA clusters
After the identification of the modular structure of TAS loci and the TLL core domain common between D. melanogaster TAS-R and TAS-L, we addressed the evolutionary origin of these sequences by studying other Drosophilidae. We performed BLAST searches using the T3 sequence against other published Drosophila genomes (FlyBase.org). In D. sechellia, we found nine scaffolds containing sequences ∼80% identical to the TLL of D. melanogaster. Scaffold 31 consists of an array of five 855-bp repeats that were identified earlier.9 Here, we found that each repeat contains two TLLs of 191 and 138 bp, and two unrelated sequences of 403 and 123 bp (Fig. 2A and Supplementary Fig. S7A). This array is flanked on one side by a TART telomeric element and on the other side by a region containing a T5 domain, a T2 domain (>90 and 85% identity, respectively) and, 8.8 kb from T2, the GM16406 gene, the ortholog of D. melanogaster Map205 the most distal gene on 3R chromosome. Scaffold 14 contains a similar 855 bp domain in proximity to the ortholog of the D. melanogaster l(2)gl gene, the most distal gene on 2L (data not shown). Using the D. sechellia 855 bp repeat, BLAST searches identified eight scaffolds within the D. simulans genome. Among them, Scaffold 3R contains the GD16184 gene, the ortholog of the Map205 gene 2.3 kb away from a region containing a T2 domain (86% identity), a T5 domain (90% identity), a TLL (>80% identity) and two related domains of 105 and 129 bp (Fig. 2B). Moreover, syntenic regions consisting of one TLL and one 129 bp-like sequence were found in the 855 bp repeats of D. sechellia, in D. simulans and in the D. melanogaster TAS-L and 0.9-kb repeat (Supplementary Fig. S7B and C). Results of the alignment between the D. sechellia 129-bp sequence and the D. melanogaster TAS-L strongly suggest that the 129-bp sequence and TLL are related (Supplementary Fig. S7B and C). In D. erecta, seven scaffolds were identified containing smaller TLL repeats (60–65 bp) 85% identical to the D. sechellia TLL. Among them, Scaffold 4820 contains four tandemly repeated domains of 998 bp which include an 128-bp sequence aligning with the TLL and which are at a distance of 13 kb from the ortholog of l(2)gl (GG24690) (Supplementary Fig. S7A). Finally, in D. yakuba, Scaffold 2L contains two almost identical TLLs of 64 bp, spaced by a region of 102 bp, 2 kb from the GE16527 gene, the ortholog of l(2)gl in this species (Supplementary Fig. S7A). Six other scaffolds in D. yakuba contain ∼170 bp 88% identical to the 274 bp internal sequence of the D. sechellia 855 bp repeats. The conserved synteny within the melanogaster subgroup suggested a telomeric location that was confirmed by in situ hybridization performed on polytene chromosomes from D. sechellia and D. simulans using a probe prepared with the 855 bp repeat of D. sechellia (Fig. 2C). When hybridization was performed on D. melanogaster chromosomes (Oregon-R-C), the 855 bp probe labelled X-TAS, 2L-TAS and 3L-TAS, as well as 3R-TAS and on the chromocenter (Fig. 2C). Finally, analysis of small RNAs extracted from D. sechellia, D. simulans and D. melanogaster ovaries revealed that the repeats identified in these species are able to produce small RNAs of a restricted size distribution of 23–29 nt with a clear uridine bias at their 5′ end (1U bias) (Fig. 2D and E) that are two key features of piRNAs.20 Therefore, these results strongly suggested that the conserved repeats identified in these species represent subtelomeric piRNA clusters.
3.4. Distribution of TAS-R family in D. melanogaster laboratory stocks
In order to address the question as to whether variability in the presence of TAS sequences exists between strains, we conducted an extensive analysis of various laboratory stocks focusing on the TAS-R family. The y1; cn1bw1sp1 genome contains at least six 3R-TAS, one 2R-TAS but no X-TAS repeats (flybase.org), while molecular analysis identified X-TAS in Lk-P(1A), NA-P(1A) and P-1152.13,19,25
We first assayed the distribution of TAS-R within the five unrelated standard laboratory stocks, i.e. Canton-S, Oregon-R-C, w1118, ywc and y1cn1bw1sp1, by performing in situ hybridization on polytene chromosomes of salivary glands of third instar larvae with a D. melanogaster X-TAS probe (Fig. 1A and Supplementary Fig. S1). This probe is expected to label specifically the three TAS-R loci due to the number of shared domains between the TAS-R family members. Control experiments using Lk-P(1A) and P-1152 lines showed strong labellings of the X and 2R telomeres for Lk-P(1A), and of the X and 3R telomeres for P-1152 (Fig. 4A and Supplementary Fig. S8). As summarized in Table 1, the five standard laboratory strains contain TAS on 2R and/or 3R autosomal loci and none on the X chromosome, except for Oregon-R-C (Supplementary Fig. S8). It is important to note that depending on individual stocks tested, Oregon-R-C possesses (this study) or lacks32 the X-TAS subtelomeric locus. In situ hybridization results for the presence/absence of X-TAS were consistent with results of PCR amplifications using specific primers for the T3 domain present in TAS-B (Table 1 and Supplementary Fig. S1), providing us with an easy assay to systematically screen for the presence/absence of X-TAS. Using sequences available in databases,1,26,28 absence of X-TAS repeats in Canton-S and y1; cn1bw1sp1 was confirmed and presence (Canton-S) or absence (y1; cn1bw1sp1) of the 0.9 kb repeats was demonstrated (Fig. 1G and H).
Figure 4.
Distribution of piRNA subtelomeric clusters in recently collected D. melanogaster. Results of analysis of the Lk-P(1A) laboratory line and three recently collected lines [Chantemesle and Reth (Europe) and Cacao H1C (South America)] are shown on first, second, third and fourth columns, respectively. (A) Polytene chromosomes of salivary gland of third instar larvae (DAPI staining) were hybridized with specific probes for TAS-R repeats. TAS-R sequences are present on the X and 2R telomeres in the Lk-P(1A) line and on X, 2R and 3R telomeres in Chantemesle, Reth and Cacao H1C. Other examples of TAS-R localization in various stocks can be found in Supplementary Figs. S8 and S9 and summarized on Tables 1 and 2. (B-G) Graphs show normalized profiles of ovarian small RNAs mapping with 0 mismatches to subtelomeric loci [T3 domain, representative of X-TAS belonging to the TAS-R family (B, C), the 0.9 kb repeats (D, E) and the TAS-L family (F, G)]. Size distribution histograms (B, D and F) and read maps (C, E and G) show the length of small RNA (20–29 nt) and the abundance of normalized piRNAs (23–29 nt) matching to each reference sequence, respectively. Numbers at the bottom right of the size distribution histograms indicate the ratio of 23–29 nt small RNAs starting with an uridine (1 U bias) (ND: Not Determined). Although the piRNA profiles between lines appear similar, differences in the amount of piRNA can be noticed. Absence of TAS-L piRNA in Lk-P(1A) suggests that the 2L and 3L-TAS loci were lost. PCR amplification screen using TAS-L specific primers identified a total of two stocks out of 43 missing the TAS-L family sequences (Tables 1 and 2). This suggested that, contrary to the TAS-R family, complete deletion of TAS-L loci does not affect viability. Other analysis of small RNA profiles can be found in Supplementary Fig. S10.
Table 1.
Distribution of TAS family in various laboratory strains
| Year of capture | T3 domaind | X-TASe | 2R-TASe | 3R-TASe | TAS-Ld | |
|---|---|---|---|---|---|---|
| Lk-P(1A)a,c | NA | + | + | + | − | − |
| NA-P(1A)a,c | NA | − | + | − | + | + |
| P-1152a,c | NA | + | + | − | + | + |
| w1118a | NA | − | − | + | + | + |
| ywa | NA | − | − | + | + | + |
| y1; cn1 bw1 sp1b | NA | − | − | + | + | + |
| Canton-Sb, OH | 1935 | − | − | + | + | + |
| Oregon-R-Cb, OR | 1925 | + | + | + | + | + |
| Grutaa, France | 1950 | − | − | + | − | + |
| BER_2b, Bermuda | 1954 | + | + | + | + | + |
| VAG_2b, Greece | 1965 | + | NT | NT | NT | + |
| Wild_11Cb, NC | 1966 | + | NT | NT | NT | + |
| Wild_5Ab, GA | 1966 | − | − | + | + | − |
| Tautavela, France | 1967 | − | − | + | − | + |
| Harwicha, MA | 1967 | − | − | + | + | + |
| Chateleta, France | 1970 | − | − | + | + | + |
| Para Wirraa, Australia | 1972 | − | − | + | + | + |
| Tachkenta, Uzbekistan | 1981 | − | − | − | + | + |
| Chimkenta, Kazakhstan | 1983 | − | NT | NT | NT | + |
aOur laboratory stocks.
bBloomington stock center.
cThese stocks were used as control for TAS.
dThe data were obtained by PCR amplification using specific primers testing the presence of T3 domain or TAS-L family.
eThe data were obtained by in situ hybridization on polytene chromosome of at least four larvae using a TAS-R probe.
NA: Not Applicable because these lines have undergone chromosomal substitutions.
In order to extend this analysis, we performed in situ hybridization screen on polytene chromosomes and/or PCR amplifications on 11 unrelated fly stocks from diverse locations in the world and introduced into our laboratory or Stock Centers several decades ago. In situ hybridization results summarized in Table 1 showed that none of the strains are totally devoid of TAS-R loci, but that the majority are devoid of X-TAS (8/11) (some examples are shown in Supplementary Fig. S8). Interestingly, BER_2, the first strain found to carry P elements in the 1950s,11 still possesses X-TAS containing P sequences (Supplementary Fig. S8). Moreover, the Para Wirra strain, initially described to contain P element sequences inserted at the 1A cytological site, suggesting the presence of X-TAS at the time,12 no longer presents X-TAS today (Table 1).
Therefore, none of the stocks tested are completely devoid of TAS-R family repeats. Nevertheless, X-TAS appear to have a particular status, since the majority of laboratory stocks tested lacks this locus (12/16), a bias that is not seen for the 2R- (1/13) or 3R-TAS (2/13) loci. This finding implicates that it may be more critical to maintain 2R- and 3R-TAS than X-TAS, suggesting a differential functional role between these TAS in fly biology.
3.5. Subtelomeric regions in D. melanogaster wild populations
To investigate the frequency of X-TAS in flies living in the wild, we generated a total of 95 D. melanogaster stocks from flies sampled in three continents (Africa, America and Europe). We performed in situ hybridizations and/or PCR amplifications immediately upon generation of all 95 stocks (i.e. on the founders of the lines) and a few months after their capture (Supplementary Fig. S9 and Supplementary Table S2). As shown in Table 2, the 23 lines tested by in situ hybridization possess at least two TAS-R loci. Lines isolated from the same location displayed polymorphism as regarding the presence of autosomic TAS within a population (see Marsais, Niort and Saul strains). In contrast to the long-term laboratory stocks, all 95 stocks established from flies recently collected from the wild possessed the X-TAS locus except for one, i.e. the line from Ilha Grande (Brazil) (Table 2 and Supplementary Table S2). In the latter case, the stock was established from one captured female. Two sublines were then generated (Ilha Grande-1 and Ilha Grande-2) that were tested 3 months post-sampling for the presence/absence of X-TAS. Since at this point one of the two sublines, Ilha Grande-1, carried no X-TAS (Table 2 and Supplementary Table S2), it is tempting to speculate that loss of the X-TAS occurred soon after this subline was established.
Finally, ovarian small RNA sequencing using three lines established from recently collected flies (Chantemesle, Reth and Cacao H1C) and three laboratory stocks (Lk-P(1A), Oregon-R-C and P-1152), all carrying X-TAS, demonstrated the production of small RNAs from the T3 domain in all cases. These small RNAs have a defined length of 23–29 nt and a 5′ uridine bias consistent with piRNA molecular properties (Fig. 4B and Supplementary Fig. S10A). Moreover, a bias toward production of sense piRNAs was noted for all stocks tested, suggesting that X-TAS repeats are asymmetric piRNA clusters. The presence of P elements (Lk-P(1A), Reth, Cacao H1C) or P-transgenes (P-1152) in X-TAS did not affect piRNA production when compared with strains devoid of P elements in X-TAS (Oregon-R-C, Chantemesle) (Fig. 4B, Supplementary Figs. S9, S10 and Supplementary Results). Interestingly, the 0.9-kb repeats were also able to produce piRNAs independent of the presence or absence of X-TAS arrays (Fig. 4D and Supplementary Fig. S10C). The 0.9-kb repeats and the X-TAS appear to be two independent piRNA clusters.
Therefore, the prevalence of functional dual-strand subtelomeric piRNA clusters in recently collected D. melanogaster stocks (22/23 stocks) is significantly higher than that in stocks kept for a long period of time in standardized laboratory conditions (4/16) (Chi square test, P < 0.001).
3.6. The X-TAS locus can be lost in stocks within 2 years of their introduction into the laboratory
The results above suggest that loss of X-TAS may occur early during establishment of stocks in the laboratory from flies collected in the wild. In order to determine the frequency of X-TAS loss upon establishment and maintenance of such fly stocks in laboratories, and estimate the kinetics of this process, two independent stocks, called A and B were established upon recent sampling of flies in the wild (Supplementary Table S2). Presence of X-TAS repeats was assayed by PCR amplifications at different time points after the sampling. After 1 year of maintenance, the stocks Saul A3-A and Saul 13-B from French Guiana have lost X-TAS repeats (Table 3 and Supplementary Table S2). We extended the analysis to strains kept for 2 years and identified seven additional stocks that no longer possess X-TAS (Niort G2-A and Niort P2.4-A; Marsais 53Ja-B, Marsais 90C-B, Niort P2.6-B, Niort L3-B and Saul FG24-B) (Table 3 and Supplementary Table S2). To confirm X-TAS loss, we further analyzed Marsais 53Ja stocks A and B 2 years after collection from the wild by in situ hybridization on polytene chromosome from salivary glands of ten male larvae (i.e. hemizygote for the X chromosome). While stock A had conserved the three TAS-R loci for all nuclei tested (n = 285) a few months post-sampling, X-TAS loci were consistently absent from all nuclei tested of stock B (n = 297) (Fig. 5). In order to increase the number of tested flies, we also individually PCR amplified 20 males from the stock B, and found that 18 out of 20 males have lost the X-TAS locus (Supplementary Table S3).
Figure 5.
Complete loss of the X-TAS locus in a two-year period. In situ hybridizations on polytene chromosomes from salivary glands from third instar larvae were performed on stocks at different time points after collection from the wild. Results are presented for two Marsais 53Ja sublines maintained separately (sublines A and B) after 2 years under laboratory conditions. (A) In stock A, the three TAS-R loci are labeled thus presenting the same pattern as when tested a few months after the capture (data not shown). Chromosomal ends of the two 3R arm homologs have been separated during the squash, showing the labeling on each of the homologous chromosomes. (B) In stock B, only two TAS (at 2R and 3R) are labeled, whereas a short time after capture all three TAS-R loci were present (data not shown) indicating the loss of X-TAS in this subline. Therefore, it is possible to witness, in the space of a two-year period, the loss of an X-TAS from a line introduced into the laboratory from the wild. Numbers in parenthesis indicate the number of nuclei studied isolated from ten male larvae. Additional in situ hybridizations are discussed in the Supplementary results.
To elucidate the structure of X chromosome extremities after X-TAS loss, PCR amplifications of X-TAS depleted stocks using specific primers for T3 (TAS-B) and the 0.9-kb repeats were performed (see above). However, although in situ hybridization with the TAS probe on the NA-P(1A) line showed a faint band on the X telomere (Supplementary Fig. S8), no PCR amplification was obtained using T3 specific primers. By designing new primer sets, we found that only the TAS-D repeat, the most divergent repeat from TAS-B, was present (Fig. 5). By using specific PCR primer sets for TAS-B, TAS-D and the 0.9-kb repeats, we found that TAS-B amplification was always associated with TAS-D and 0.9-kb repeat PCR amplifications, and that TAS-D amplification was always associated with 0.9-kb repeats amplification. Therefore, the organization of X subtelomeres, when present, is similar to the one described from the minichromosome more than 20 years ago. Figure 3 displays a schematic representation of subtelomeric structures as deduced from the PCR amplifications. It shows that three of the recently collected lines that lost X-TAS still possess the TAS-D repeat like NA-P(1A) (Fig. 3B), another one of these lines that has retained the 0.9-kb repeats like Canton-S (Fig. 3C) and five other lines that have lost the three domains tested (TAS-B, TAS-D and 0.9 kb) like y1; cn1bw1sp1 (Fig. 3D). Finally, we also tested strains, which have been maintained in laboratory conditions for decades and are devoid of X-TAS locus. Seven out of the 14 lines tested have a deletion of the whole X subtelomere (Fig. 3D) and three have only retained the 0.9-kb repeats (Fig. 3C).
What is the molecular mechanism of TAS loss? Anderson et al.33 suggested the existence of a recombination hot spot in the most distal portion of the X chromosome consistent with a role in evolution of subtelomeric DNA. Moreover, Novitski34 proposed the existence of a meiotic drive in the Drosophila oocyte that favors transmission of shorter telomeres. Our results suggest that loss of the three types of repeats (i.e. TAS-B, TAS-D and the 0.9 kb repeats) might happen after terminal chromosomal breakage rather than by random internal deletion in the locus. These events could then have been selected for meiotic drive that would confer a segregation advantage for flies raised under laboratory conditions. Since males are hemizygotes for the X chromosome, this loss could then be rapidly fixed in laboratory populations. In any case, we assume that these terminal deletions were subsequently healed by retrotransposition of telomeric HTT elements as it has been previously described.25,35
4. Discussion
4.1. Subtelomeres in Drosophila are ancestral piRNA clusters
Our investigations reveal that not only members of the TAS-R family are more related to each other than previously thought, but that there is also a relation between TAS-R and TAS-L. Besides the three tandemly repeated INV-4 solo LTRs already known to define the TAS-R family, we identified five other shared domains (named T1, T2, T3, T4 and T5). The T3 domain was found to be restricted to X-TAS, along with another sequence, newly characterized here, of a 158-bp that we called TLL. Indeed, in silico analyses demonstrated that TLLs are the tandemly repeated unit substructure of TAS-L. Interestingly, specific polymorphisms exist among the X-, 2R-, 3R-, 2L- and 3L-TAS loci, suggesting the existence of a mechanism(s) that prevents exchange in trans between subtelomeric loci in D. melanogaster, contrary to what has been shown for humans and S. cerevisiae.36 This is consistent with previous results showing suppression of crossing-over in regions proximal to telomeres in Drosophila.37
In silico comparisons show that TLL sequences are found in related members of the melanogaster subgroup, but not in more distantly related Drosophilidae. Our results indicate that a TLL duplication event, giving rise to a 129-bp sequence, occurred after the divergence between D. melanogaster/D. sechellia/D. simulans and D. erecta/D. yakuba (see model in Fig. 6). In agreement with the results presented here, we have hypothesized that TLL and the 129-bp sequence are thus paralogs and along with T2 and T5 were already existed in a common ancestor (Fig. 6). According to this hypothesis, 0.9-kb repeats and the TAS-R and TAS-L families could have emerged then from successive genomic events (deletion, insertion and duplication) in D. melanogaster (Fig. 6). Finally, results of in situ hybridization and ovarian small RNA sequencing experiments strongly suggest that the TLL sequence could be at the origin of subtelomeric dual-strand piRNA clusters in the melanogaster subgroup that might be involved in TE repression.
Figure 6.
Model of succession of genomic events during the course of TAS family repeat evolution in the melanogaster subgroup. Shown on the melanogaster subgroup phylogenetic tree, an ancestral TLL gave rise to subtelomeric repeats in D. erecta and D. yakuba on the one hand, while on the other hand duplication of TLL and insertion of sequences leading to the T2 and T5 domains occurred in the common ancestor of D. sechellia, D. simulans and D. melanogaster. During speciation of D. melanogaster, deletion of the T2 and T5 domains followed by amplification events led to the establishment of TAS-L and the 0.9 kb repeats. The TAS-R family arose by insertion of sequences leading to the T1, T4, T3′ and T3″ domains. The T3′, T3″ and TLL correspond to the full-length of the T3 domain. More recently, an INV-4 TE inserted into the locus, recombined so as to leave a solo LTR and then was amplified. A scenario of successive events (insertion, deletion and recombination) giving rise to the three TAS-R family members known today is proposed here. Note that domains and repeat units are not drawn to scale to facilitate the graphic representation. The origin of the ‘T’ domains is unknown and discussed in the text. Telomeres are to the left of the structural scheme for each chromosomal region depicted.
This X subtelomeric region was particularly informative in this study since this region appears to be at the crossroad between the two main TAS families of D. melanogaster autosomes: X-TAS possess large regions of homology with the TAS-R family, via the T1, T2, T3, T4, T5 and INV-4 sequences, as well as with the TAS-L family, via TLL and the 129-bp sequences present in the adjacent 0.9-kb repeats (Fig. 1B and Supplementary Fig. S7C). Interestingly, it was not possible to find an origin for the different “T” domains. Therefore, it is tempting to speculate that TAS derive from successive ancestral insertions of diverse TEs that got trapped, similar to the recently trapped INV-4 and P elements. According to this scenario, the majority of TAS would consist of remnants of extinct TEs.
4.2. Short-term dynamics of subtelomeric piRNA clusters
Subtelomeric variability, i.e. variability in the absence/presence of specific sequence elements and/or in the number of repeats, has been described previously,6,33,35,38 however, the dynamics of subtelomeric sequence element gain or loss has not been addressed to a great extent, nor has the potential impact of subtelomeric dynamics on strain fitness.
Here, we report an original survey of the genetic variation at X subtelomeres between D. melanogaster laboratory stocks and recently collected stocks. From this study, no laboratory stock or wild-type stock completely devoid of TAS-R was identified, whereas TAS-L were altogether absent in some stocks. This suggests that TAS-L, but not TAS-R, are dispensable in some cases. In addition, the majority of laboratory stocks were found to be devoid of an X-TAS locus. In contrast, X-TAS repeats were present in flies directly sampled from the wild and in recently established stocks from flies collected in Europe, South America and Africa. However, once these recently established stocks were maintained under standardized laboratory conditions for several months up to 2 years and retested X-TAS were no longer present for some of the sublines. Therefore, we were able to follow X-TAS loss in real time, most likely due to frequent chromosomal breakage rather than internal deletions inside the locus (Fig. 3). Our data suggest that loss of X-TAS is not influenced by the absence or presence of P element insertions in this locus (Supplementary results), nor by temperature (see stocks A and B maintained at 18 and 25°C, respectively) (Supplementary Table S2), nor by the geographical origin of the flies. Therefore, we hypothesize that X-TAS fulfill a fundamental function for life in the wild that is not required under laboratory conditions. Alternatively, X-TAS loss under laboratory conditions could be simply the consequence of inbreeding effects due to isogenisation.
The nature of the selective forces acting on subtelomeric polymorphism identified here remains unclear. Nevertheless, subtelomeric piRNA clusters, especially X-TAS, appear to have an impact on fitness in the natural environment while not affecting the viability of flies raised under laboratory conditions. To our knowledge, this is the first systematic study monitoring natural loss of a piRNA cluster under specific conditions. The apparent paradox in the fact that loci involved in genome defense are subject to an absence/presence polymorphism suggests possible functional redundancy between the 140 piRNA clusters scattered throughout the D. melanogaster genome. This may thus provide an efficient genomic mechanism to control a tremendous load of TEs in gonads. Therefore, piRNA clusters appear to be capable of rapid evolution may be because they are, on the one hand, able to attract several kb of endogenous and/or exogenous TE copies and, on the other hand, able to lose sequences that are several kb in length without affecting viability under some, still undetermined environmental conditions. This capacity offers a time window for TEs to be derepressed (as it was observed for P elements in the Marsais 53Ja B, Supplementary Fig. S9). This opportunity for transposition appears to be essential to shape the genome by allowing new insertions that generate genomic diversity, as well as novelty. Among these new insertions, rare cases will be maintained in particular environments, as in the case of telomeres in Drosophila, CENP-B of centromeres and the RAG system in vertebrates.39
Supplementary Material
Acknowledgements
We thank Clément Carré, Emile Civel, Aurélie Hua-Van, Claude Lespinet, Gilbert and Micheline Lespinet, Pauline Marie, Valerie Ribeiro, Claude et Michèle Teysset, and Anne Laure Todeschini for their help in collecting flies, the Bloomington Stock Center for providing stocks, FlyBase.org for providing valuable databases and ARTbio/Bioinformatics analyses platform at the IBPS for the development of valuable tools. We thank Jean-François Gilles and Susanne Bolte for the help with confocal microscopy carried out at the Institute of Biology Paris-Seine Imaging Facility, supported by the “Conseil Regional Ile-de France”, the French national research council (CNRS) and Sorbonne University, UPMC Univ Paris 06. We thank Ritha Zamy and Shang Nong HU for technical assistance, Robert W. Levis for helpful discussions and Jean-Michel Gibert, Elena Casacuberta, Antoine Boivin, Sandra Duharcourt and Anne-Marie Pret for critical reading of the manuscript.
Conflict of interest
None declared.
Supplementary data
Supplementary data are available at DNARES Online.
Funding
This study was supported by fellowships from the Ministère de l’Enseignement Supérieur et de la Recherche to A.A.-L. and by grants from the Association de la Recherche contre le Cancer (Fondation ARC, SFI20121205921, SFI20131200470), the Fondation pour la Recherche Médicale, (FRM DEP20131128532), the Agence Nationale de la Recherche (ANR, project “plastisipi”) to S.R., and the University Pierre et Marie Curie (Emergence EME1223) to L.T. Sampling of D. melanogaster in French Guiana was funded by a CEBA grant (MONISPEC ANR-10-LABX-25-01) and a CNRS Nouragues Travel Grant 2015 (ACCISPEC NTG 2015) to W.J.M. The funders had no role in study design, data collection, analysis, decision to publish, or preparation of the manuscript.
References
- 1. Karpen G.H., Spradling A.C.. 1992, Analysis of subtelomeric heterochromatin in the Drosophila minichromosome Dp1187 by single P element insertional mutagenesis. Genetics, 132, 737–753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Levis R.W., Ganesan R., Houtchens K., Tolar L.A., Sheen F.M.. 1993, Transposons in place of telomeric repeats at a Drosophila telomere. Cell, 75, 1083–1093. [DOI] [PubMed] [Google Scholar]
- 3. Walter M.F., Jang C., Kasravi B., et al. 1995, DNA organization and polymorphism of a wild-type Drosophila telomere region. Chromosoma, 104, 229–241. [DOI] [PubMed] [Google Scholar]
- 4. Abad J.P., De Pablos B., Osoegawa K., De Jong P.J., Martin-Gallardo A., Villasante A.. 2004, Genomic analysis of Drosophila melanogaster telomeres: full-length copies of HeT-A and TART elements at telomeres. Mol. Biol. Evol., 21, 1613–1619. [DOI] [PubMed] [Google Scholar]
- 5. Bergman C.M., Quesneville H., Anxolabéhère D., Ashburner M.. 2006, Recurrent insertion and duplication generate networks of transposable element sequences in the Drosophila melanogaster genome. Genome Biol., 7, R112. [DOI] [PMC free article] [PubMed]
- 6. Levis R.W. 1989, Viable deletions of a telomere from a Drosophila chromosome. Cell, 58, 791–801. [DOI] [PubMed] [Google Scholar]
- 7. Yin H., Lin H.. 2007, An epigenetic activation role of Piwi and a Piwi-associated piRNA in Drosophila melanogaster. Nature, 450, 304–308. [DOI] [PubMed] [Google Scholar]
- 8. Silva-Sousa R., Casacuberta E.. 2013, The JIL-1 kinase affects telomere expression in the different telomere domains of Drosophila. PLoS One, 8, e81543.24244743 [Google Scholar]
- 9. Mason J.M., Villasante A.. 2014, Subtelomeres in Drosophila and other Diptera.InSubtelomeres, Louis E. J., Becker M .M. (eds.), Springer: Berlin, 211–225. [Google Scholar]
- 10. Huisinga K.L., Riddle N.C., Leung W., et al. 2016, Targeting of P-element reporters to heterochromatic domains by transposable element 1360 in Drosophila melanogaster. Genetics, 202, 565–582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Anxolabehere D., Kidwell M.G., Periquet G.. 1988, Molecular characteristics of diverse populations are consistent with the hypothesis of a recent invasion of Drosophila melanogaster by mobile P elements. Mol. Biol. Evol., 5, 252–269. [DOI] [PubMed] [Google Scholar]
- 12. Ronsseray S., Lehmann M.,, Anxolabehere D.. 1989, Copy number and distribution of P and I mobile elements in Drosophila melanogaster populations. Chromosoma, 98, 207–214. [DOI] [PubMed] [Google Scholar]
- 13. Ronsseray S., Lehmann M., Anxolabehere D.. 1991, The maternally inherited regulation of P elements in Drosophila melanogaster can be elicited by two P copies at cytological site 1A on the X chromosome. Genetics, 129, 501–512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Josse T., Teysset L., Todeschini A. L., Sidor C. M., Anxolabehere D., Ronsseray S.. 2007, Telomeric trans-silencing: an epigenetic repression combining RNA silencing and heterochromatin formation. PLoS Genet., 3, 1633–1643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Roche S.E., Rio D.C.. 1998, Trans-silencing by P elements inserted in subtelomeric heterochromatin involves the Drosophila Polycomb group gene, Enhancer of zeste. Genetics, 149, 1839–1855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Ronsseray S., Josse T., Boivin A.,, Anxolabehere D.. 2003, Telomeric transgenes and trans-silencing in Drosophila. Genetica, 117, 327–335. [DOI] [PubMed] [Google Scholar]
- 17. Brennecke J., Aravin A.A., Stark A., et al. 2007, Discrete small RNA-generating loci as master regulators of transposon activity in Drosophila. Cell, 128, 1089–1103. [DOI] [PubMed] [Google Scholar]
- 18. Czech B., Hannon G.J.. 2016, One loop to rule them all: the ping-pong cycle and piRNA-guided silencing. Trends Biochem. Sci, 41, 324–337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Josse T., Maurel-Zaffran C., de Vanssay A., et al. 2008, Telomeric trans-silencing in Drosophila melanogaster: tissue specificity, development and functional interactions between non-homologous telomeres. PLoS One, 3, e3249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Brennecke J., Malone C.D., Aravin A.A., Sachidanandam R., Stark A.,, Hannon G.J.. 2008, An epigenetic role for maternally inherited piRNAs in transposon silencing. Science, 322, 1387–1392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Todeschini A.L., Teysset L., Delmarre V.,, Ronsseray S.. 2010, The epigenetic trans-silencing effect in Drosophila involves maternally-transmitted small RNAs whose production depends on the piRNA pathway and HP1. PLoS One, 5, e11032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. de Vanssay A., Bouge A.L., Boivin A., et al. 2012, Paramutation in Drosophila linked to emergence of a piRNA-producing locus. Nature, 490, 112–115. [DOI] [PubMed] [Google Scholar]
- 23. Muerdter F., Olovnikov I., Molaro A., et al. 2012, Production of artificial piRNAs in flies and mice. RNA, 18, 42–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. O’Kane C.J., Gehring W.J.. 1987, Detection in situ of genomic regulatory elements in Drosophila. Proc. Natl Acad. Sci. U. S. A., 84, 9123–9127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Marin L., Lehmann M., Nouaud D., Izaabel H., Anxolabehere D., Ronsseray S.. 2000, P-Element repression in Drosophila melanogaster by a naturally occurring defective telomeric P copy. Genetics, 155, 1841–1854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Benos P.V., Gatt M.K., Murphy L., et al. 2001, From first base: the sequence of the tip of the X chromosome of Drosophila melanogaster, a comparison of two sequencing strategies. Genome Res., 11, 710–730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Boussy I.A., Itoh M.. 2004, Wanderings of hobo: a transposon in Drosophila melanogaster and its close relatives. Genetica, 120, 125–136. [DOI] [PubMed] [Google Scholar]
- 28. Adams M.D., Celniker S.E., Holt R.A., et al. 2000, The genome sequence of Drosophila melanogaster. Science, 287, 2185–2195. [DOI] [PubMed] [Google Scholar]
- 29. Gouy M., Guindon S.,, Gascuel O.. 2010, SeaView version 4: a multiplatform graphical user interface for sequence alignment and phylogenetic tree building. Mol. Biol. Evol., 27, 221–224. [DOI] [PubMed] [Google Scholar]
- 30. Hermant C., Boivin A., Teysset L., et al. 2015, Paramutation in Drosophila requires both nuclear and cytoplasmic actors of the piRNA pathway and induces Cis-spreading of piRNA production. Genetics, 201, 1381–1396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Langmead B., Trapnell C., Pop M.,, Salzberg S.L.. 2009, Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol., 10, R25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Andreyeva E.N., Belyaeva E.S., Semeshin V.F., Pokholkova G.V.,, Zhimulev I.F.. 2005, Three distinct chromatin domains in telomere ends of polytene chromosomes in Drosophila melanogaster Tel mutants. J. Cell Sci., 118, 5465–5477. [DOI] [PubMed] [Google Scholar]
- 33. Anderson J.A., Song Y.S.,, Langley C.H.. 2008, Molecular population genetics of Drosophila subtelomeric DNA. Genetics, 178, 477–487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Novitski E. 1951, Non-random disjunction in Drosophila. Genetics, 36, 267–280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Kern A.D.,, Begun D.J.. 2008, Recurrent deletion and gene presence/absence polymorphism: telomere dynamics dominate evolution at the tip of 3L in Drosophila melanogaster and D. simulans. Genetics, 179, 1021–1027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Mefford H.C., Trask B.J.. 2002, The complex structure and dynamic evolution of human subtelomeres. Nat. Rev. Genet., 3, 91–102. [DOI] [PubMed] [Google Scholar]
- 37. Lindsley D.L., Sandler L.. 1977, The genetic analysis of meiosis in female Drosophila melanogaster. Philos. Trans. R. Soc. Lond. B Biol. Sci., 277, 295–312. [DOI] [PubMed] [Google Scholar]
- 38. Mechler B.M., McGinnis W.,, Gehring W.J.. 1985, Molecular cloning of lethal(2)giant larvae, a recessive oncogene of Drosophila melanogaster. EMBO J., 4, 1551–1557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Volff J.N. 2006, Turning junk into gold: domestication of transposable elements and the creation of new genes in eukaryotes. Bioessays, 28, 913–922. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.