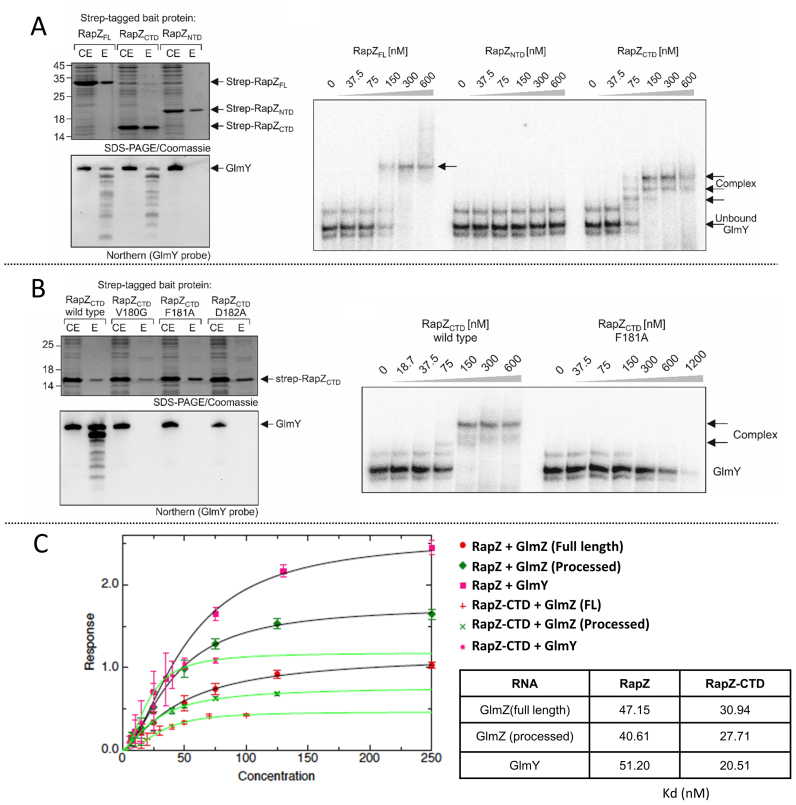
Figure 7.
Assessment of the role of individual domains of RapZ in RNA binding. (A) RNA binding activity is associated with the RapZ-CTD but not with the NTD. Full-length Strep-RapZ, Strep-RapZ-CTD and Strep-RapZ-NTD were purified via StrepTactin affinity chromatography from strain Z903 lacking endogenous rapZ. Aliquots of the elution fractions (E) were analyzed by 15% SDS-PAGE/Coomassie staining (top) and in parallel used to extract RNA, which was subsequently analyzed by northern blotting using a probe specific for GlmY (bottom). The amounts of RNA loaded were adjusted to equal protein concentrations, as determined by the SDS-PAGE gel (top). In addition, the cell extracts (CE) of the various strains harvested prior to protein purification were analyzed alongside the elution fractions. Right: EMSA experiments using purified proteins and radio-labelled GlmY. (B) Wild-type Strep-RapZ-CTD or mutants V180G, F181A and D182A were purified by StrepTactin affinity chromatography from strain Z903 and analysed as in (A). Right: EMSA experiment showing that RapZ CTD harbouring the F181A mutation is unable to bind radiolabelled GlmY in vitro. (C) RapZ-CTD binds RNA targets with higher affinity than the full-length protein. Dissociation constants were calculated for full-length RapZ (1–284) or RapZ-CTD (154–284) by biolayer interferometry using an Octet Red 96 device. The measurements were performed by fitting the response from three independent experiments, and error bars represent standard deviations from the mean.