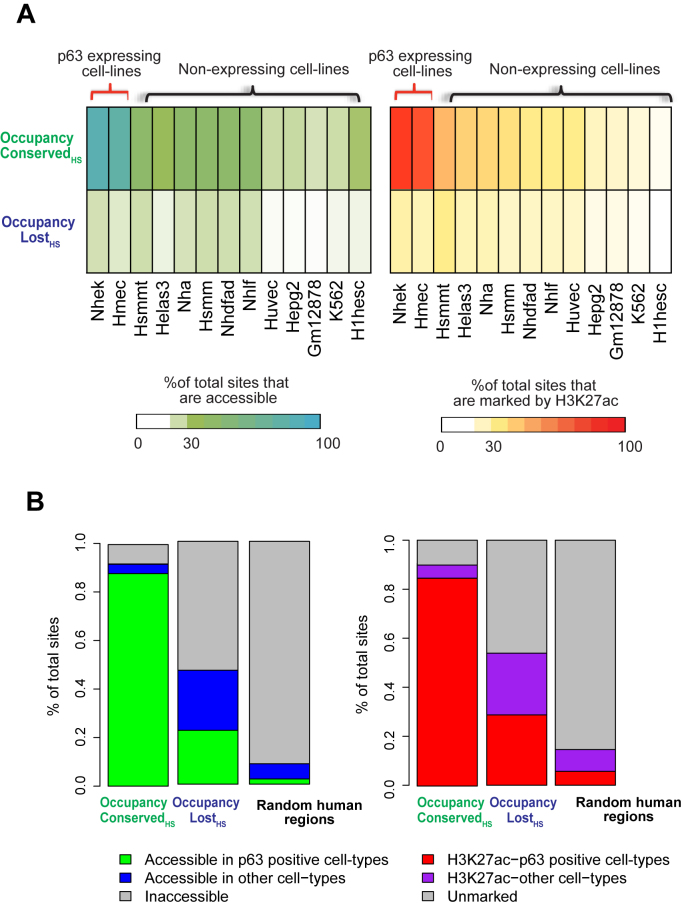
Figure 6.
Occupancy Lost mouse p63 targets might be functional in other contexts in human development. Occupancy Conserved and Occupancy Lost p63 targets are mapped to their homologous regions in humans. These genomic locations are overlapped with DNase-Seq and H3K27ac peaks in 13 diverse human cell-types, as generated by the ENCODE consortium. (A) Heatmaps: left panel: represents the percentage of genomic sites that overlap with DNase-Seq peaks in each individual cell-type. Right panel: represents the percentage of genomic sites that overlap with H3K27ac peaks in each individual cell-type. (B) Stacked bar plots: left panel: represents percentage of genomic sites that are accessible (at least 1 bp overlap with DNase-Seq peaks) across any cell-type, right panel: represents percentage of genomic sites that are marked by H3K27ac (at least 1 bp overlap with H3K27ac peaks) in any cell-type. Regions that are accessible or marked by H3K27ac in p63 negative cell-lines are colored separately from the rest. Random human genomic regions serve as negative control.