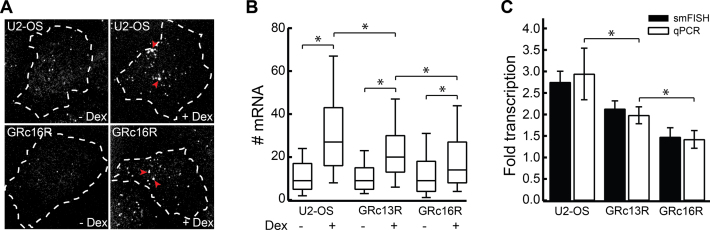
Figure 3.
GRc13R and GRc16R repress SGK1 transcription. (A) smFISH of SGK1 mRNA in wild-type U2-OS cells (upper panels) and U2-OS cells expressing GRc16R (lower panels) in the absence (left panels) and presence (right panels) of 1 μM Dex. Dashed lines indicate the nuclear membrane. Arrowheads highlight loci of nascent transcription. The contrast was adjusted individually for each panel. (B) mRNA distributions in wild-type U2-OS cells and U2-OS cells expressing GRc13R and GRc16R in absence and presence of 1 μM Dex. (*) indicates a significant difference according to the Wilcoxon–Mann–Whitney two-sample rank test, two tailed, P < 0.05. (C) Fold change of transcription calculated as ratio between the mean number of mRNA molecules upon Dex induction and the mean number of mRNA molecules in absence of Dex for wild-type U2-OS cells (308/304 cells) and U2-OS cells expressing GRc13R (308/294 cells) and (GRc16R (270/211 cells), or from qPCR experiments (triplicates on at least three biological replicates). (*) indicates a significant difference according to Student's t-test, two-tailed, P < 0.05. Error bars denote s.e.m. (smFISH) or s.d. (qPCR).