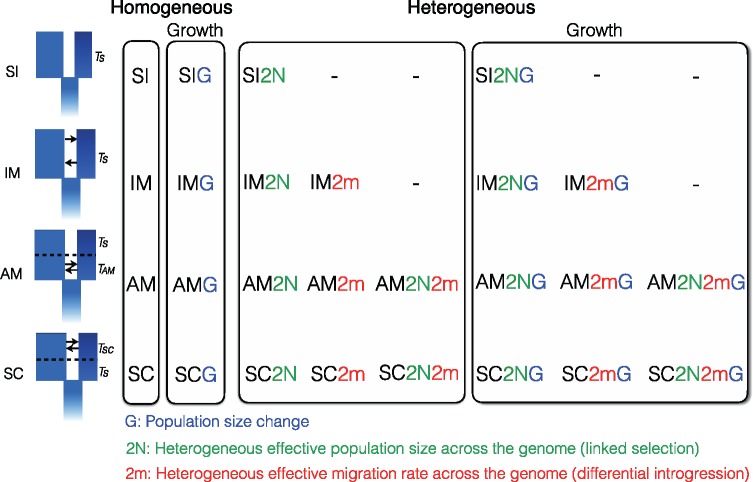
Fig. 2.
—The 26 models implemented in this study. The models implemented here represent extensions of the four classical models of divergence: “Strict Isolation” (-SI), “Isolation with Migration” (-IM), “Ancient Migration” (-AM) and “Secondary Contact” (-SC). Briefly, TS corresponds to the duration of complete isolation between diverging populations and TAM and TSC correspond to the duration of gene-flow in AM and SC models, respectively. The first extension of these four models accounts for temporal variation in effective populations size (-G models), allowing independent expansion/contraction of the diverging populations. The last categories correspond to “Heterogeneous gene flow” models, which integrate parameters allowing genomic variations in effective migration rate (-2 m), effective population size (-2 N) or both (-2m2N) to account for genetic barriers and selection at linked sites.