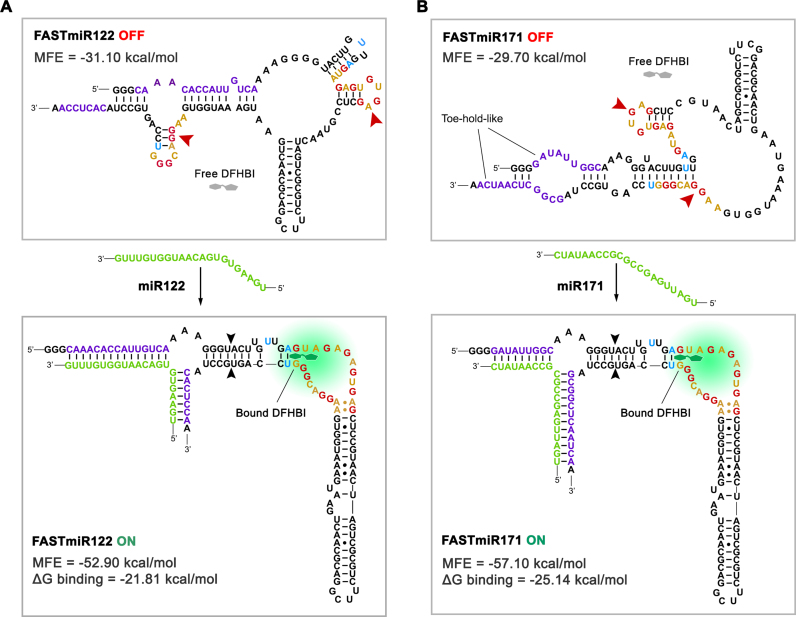
Figure 2.
OFF- and ON-states of FASTmiR122 and FASTmiR171. (A) The predicted structure of the FASTmiR122 OFF-state and the ON-state with the miR122 target. The OFF-state contains a disrupted base triple and G-quadruplex that forms two small hairpin structures (arrowheads). Free DFHBI (gray) cannot bind and then fluoresce. The miR122 (green) binds and opens the structure, resulting in a re-formed modSpinach (right of black arrowheads) that can bind DFHBI and then fluorescence. The FASTmiR122 ON-state bound by miR122 is favored thermodynamically (MFE = −52.90 kcal/mol) compared to the unbound OFF-state (MFE = −31.10 kcal/mol. ΔG binding =−21.81 kcal/mol). (B) The predicted structure of the FASTmiR171 OFF-state and the ON-state with the miR171 target. The G-quadruplex of the OFF-state is disrupted by one hairpin structure and one paired region (arrowheads). The OFF sequence has two toe-hold-like regions where the miR171 (green) can enter, bind and open the OFF-state structure. The FASTmiR sensor then switches to the ON-state that has a reformed modSpinach (right of arrowhead) that can fluoresce. The FASTmiR171 ON-state bound by miR171 is favored thermodynamically (MFE = 57.10 kcal/mol) compared to the unbound OFF-state (MFE =−29.7 kcal/mol. ΔG binding= −25.14 kcal/mol). The OFF-states in A and B are drawn based on predicted structure from Vienna RNA secondary structure server. MFE and ΔG binding were calculated using Vienna RNAfold and RNAcofold (version 2.1.1). The ON-states in A and B are drawn according to Huang et al. (18).