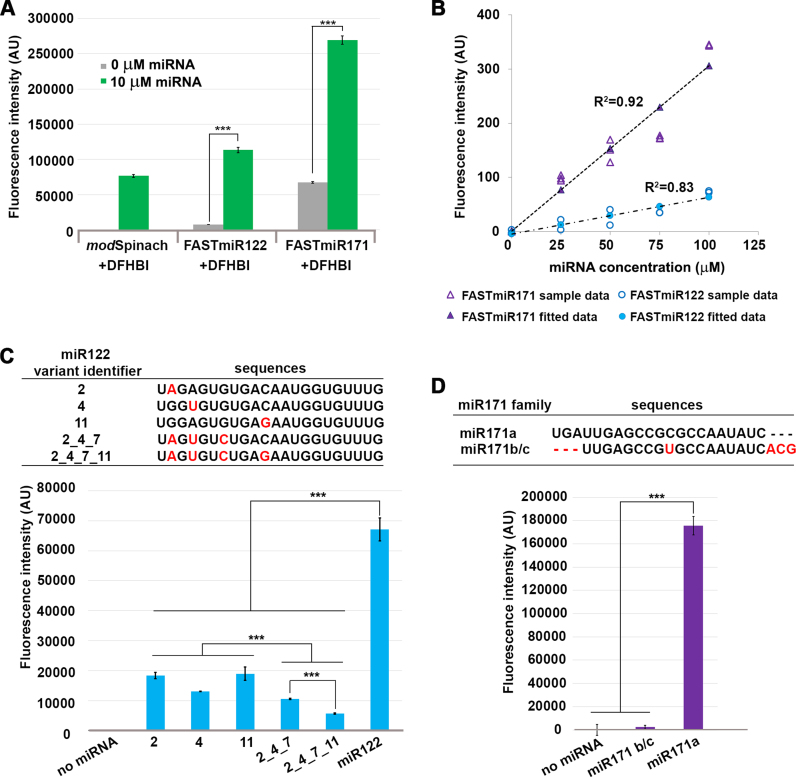
Figure 3.
Characterization of FASTmiR122 and FASTmiR171 sensors in vitro. (A) Fluorescence intensity of FASTmiR122 and FASTmiR171 with 0 or 10 uM of cognate miRNAs. FASTmiR122 with miR122 show fluorescence equivalent to the modSpinach positive control. FASTmiR171 with miR171 showed an increased fluorescence compared to the 0 mM miR171 negative control, but less than the modSpinach positive control. Fluorescence values were normalized by subtracting background DFHBI fluorescence in the water control (number of considered samples, n = 4). (B) Dose-response curves of FASTmiR122 and FASTmiR171. Fluorescence intensity increased linearly with increasing concentrations of miRNAs (0, 25, 50, 75 and 100 μM miR122 (or miR171). Fluorescence values were normalized to the sensor alone background DFHBI fluorescence. (C and D) Specificity of the FASTmiR122 and FASTmiR171 sensors to their cognate miRNAs. FASTmiR122 showed significantly higher fluorescence when incubating with miR122, compared to miR122 variant with 1–4 point mutations. FASTmiR171 showed significantly higher fluorescence when incubated with its specific miR171 target, miR171a compared to its naturally occurring family member miR171b/c or no miRNA. Fluorescence values were normalized by subtracting background DFHBI fluorescence in the water control (n = 4). Significance codes: ‘***’ for <0.001, ‘**’ for <0.01, ‘*’ for < 0.05.