Abstract
Human type II topoisomerase (Top2) isoforms, hTop2α and hTop2β, are targeted by some of the most successful anticancer drugs. These drugs induce Top2-mediated DNA cleavage to trigger cell-death pathways. The potency of these drugs correlates positively with their efficacy in stabilizing the enzyme-mediated DNA breaks. Structural analysis of hTop2α and hTop2β revealed the presence of methionine residues in the drug-binding pocket, we therefore tested whether a tighter Top2-drug association may be accomplished by introducing a methionine-reactive Pt2+ into a drug to further stabilize the DNA break. Herein, we synthesized an organoplatinum compound, etoplatin-N2β, by replacing the methionine-juxtaposing group of the drug etoposide with a cis-dichlorodiammineplatinum(II) moiety. Compared to etoposide, etoplatin-N2β more potently inhibits both human Top2s. While the DNA breaks arrested by etoposide can be rejoined, those captured by etoplatin-N2β are practically irreversible. Crystallographic analyses of hTop2β complexed with DNA and etoplatin-N2β demonstrate coordinate bond formation between Pt2+ and a flanking methionine. Notably, this stable coordinate tether can be loosened by disrupting the structural integrity of drug-binding pocket, suggesting that Pt2+ coordination chemistry may allow for the development of potent inhibitors with protein conformation-dependent reversibility. This approach may be exploited to achieve isoform-specific targeting of human Top2s.
INTRODUCTION
Imbalances in enzyme activity are etiological factors for numerous diseases, including inflammation, metabolic disorders, cardiovascular irregularities and cancers. Therefore, modulation of enzyme function by bioactive small molecules is a commonly employed therapeutic strategy and many successful drugs are enzyme inhibitors or poisons (1). The majority of drugs bind their targets via non-covalent forces, rendering the interactions reversible in nature. In contrast, irreversible inhibition has been achieved mainly via covalent bond formation between an inhibitor and its target (2,3). Despite the superior in vitro potency displayed by these so-called covalent inhibitors, their broader clinical applications are usually limited by pronounced adverse effects due to off-target reactivity and potential immunogenicity arising from the resulting protein-inhibitor adducts (4–6). Knowing that the stability of coordination complexes is determined in part by the number and geometric distribution of metal-coordinating ligands (7), we envisioned that the coordination bond formed between a transition metal ion incorporated in an organic scaffold and reactive side chain functional group(s) in a target protein may exhibit conditional lability. Perturbing the conformational state of the target protein may alter the spatial arrangement of ligands, leading to rupture of the coordination linkage. Here, we performed structure-based development of an organoplatinum compound capable of inducing type II topoisomerase (Top2)-mediated DNA breaks to show that the generally irreversible and thus highly efficient enzyme-targeting based on Pt2+ coordination chemistry may become dissociable upon protein unfolding, which illustrates a potential benefit of employing metal coordination chemistry in drug development.
Top2 resolves topological entanglements in DNA by first catalyzing the formation of a transient covalent enzyme–DNA adduct termed Top2 cleavage complex (Top2cc), which harbors a double-strand DNA break to allow for the following passage of another DNA segment (8,9). A number of clinically active anticancer drugs arrest the transiently formed Top2cc to exploit its latent yet lethal cytotoxicity; subsequent collision between the trapped Top2cc and DNA-tracking activities generates a bulky DNA lesion and in turn causes cell death (10–13). Structural analyses revealed drug intercalation between the base pairs flanking the DNA cleavage site, which effectively stabilizes Top2cc by blocking religation of the cleaved DNA ends (14–18). The specificity displayed by these drugs toward the Top2-induced DNA cleavage site can be rationalized by their interactions with the surrounding protein residues. The presence of methionine residue(s) in the drug-binding pocket in the two human Top2 isoforms (14,16,19), hTop2α and hTop2β, suggest that site-specific incorporation of a Pt2+ reactive center into a drug may enable the formation of a Pt2+–thioether bond with the methionine side chain and boost the drug’s efficacy by strengthening its interaction with Top2cc.
MATERIALS AND METHODS
Chemical synthesis of etoplatins
Refer to Supporting Online Material for a detailed description of the synthetic scheme (Supplementary Schemes S1–3).
Construction, expression and purification of recombinant proteins
The coding sequence of hTop2αcore (residues 429–1188, the DNA binding and cleavage core of hTop2α) was sub-cloned into pET51b (Novagen) from the YEpWob6 plasmid (20). The resultant plasmid (named 51bDBCCα), as well as the 51bDBCCβ plasmid (harbors the coding sequence of hTop2βcore (residues 445–1201, the DNA binding and cleavage core of hTop2β)), was transformed to Escherichia coli BL21 (DE3) Star for protein expression. The constructed YEpWob6 and YEphTOP2B plasmids (21,22) respectively harboring the coding sequences of hTop2α△CTD (residues 29–1221) and hTop2β△CTD (residues 45–1201) were delivered to yeast BCY123 cells for protein expression. The primer sequences for molecular cloning and further details of recombinant protein purification and preparation are listed in the Supplementary Material and Methods.
Crystallization and post-crystallization drug replacement
Crystallization procedures used for preparing both hTop2αcore–DNA–etoposide and hTop2βcore–DNA–etoposide ternary complexes have been described in our previous work (14). Briefly, the protein solution for crystallization is composed of purified hTop2αcore/hTop2βcore (5 mg/ml, ∼28 μM), 1 mM etoposide (diluted from 50 mM stock in pure dimethyl sulfoxide (DMSO)) and ∼34 μM of the 20 bp dsDNA substrate (5′-AGCCGAGCTGCAGCTCGGCT-3′) in gel filtration buffer (50 mM Tris–HCl (pH 7.0), 200 mM KCl, 5 mM MnCl2, 1 mM ethylenediaminetetraacetic acid (EDTA), 2 mM β-mercaptoethanol). Then, 1 μl of the protein solution was mixed with an equal amount of the reservoir solution (100 mM magnesium acetate, 50 mM 2-(N-morpholino)ethanesulfonic acid pH 5.6 and 26% 2-methyl-2,4-pentanediol (MPD)) and equilibrated against 200 μl of the reservoir solution using the hanging-drop vapor diffusion method with VDX™ plate (Hampton Research) at 4°C. Single crystals suitable for data collection usually appear within 2 weeks. Crystals were harvested by transferring into mother liquor containing 30% MPD before looping and flash-freezing in liquid nitrogen for data collection.
We followed the previously described procedure for post-crystallization drug replacement (16) to obtain the hTop2βcore cleavage complex stabilized by etoplatin-N2β and etoplatin-N2α. Briefly, etoposide was soaked out by transferring the hTop2βcore–DNA–etoposide crystal into mother liquor containing 30% MPD at 4°C for 16 h. Etoplatin-N2β and -N2α were then soaked in by adding 1 mM of respective drug (dissolved in pure DMSO) to the drop containing drug-free crystals at 4°C for 16 h. All of the crystals were harvested by transferring into mother liquor containing 30% MPD before looping and flash-freezing in liquid nitrogen for data collection. To test the stability/reversibility of the resultant hTop2βcore–DNA–etoplatin–N2β and hTop2βcore–DNA–etoplatin–N2α complexes, these etoplatin-bound crystals were subjected to two additional soaking steps before being harvested for data collection: first in drug-free mother liquor (16 h at 4°C) followed by a second soaking (1–7 days at 4°C) in mother liquor containing another hTop2-targeting drug mitoxantrone (1 mM).
Data collection and structure determination
All diffraction datasets were collected at National Synchrotron Radiation Research Center (NSRRC), Taiwan (beamlines BL13B1 and BL15A1) and processed using the HKL2000 program suite (23). The structure of hTop2αcore–DNA–etoposide was solved by molecular replacement (MR) with phenix.automr using the hTop2βcore–DNA complex taken from the hTop2βcore–DNA–etoposide structure (PDB ID: 3QX3) (14) as a search model followed by model building with phenix.autobuild (24). One cleavage complex was found per asymmetric unit. The bound drugs were built into the Fo–Fc density maps manually using Coot (25), the structure then underwent rounds of manual model rebuilding and refinement with Coot and phenix.refine. The structures of hTop2βcore–DNA–etoplatin-N2β and hTop2βcore–DNA–etoplatin-N2α were also solved by MR using the program and search model mentioned above. After subjecting the MR solutions to rigid body and positional refinement with phenix.refine, the three main components of etoplatins (the aglycone core, E-ring and dichlorodiammineplatinum(II) moiety; see Figure 1) can be unambiguously placed in the resulting Fo–Fc maps. For the etoplatin-N2β-bound structure, continuous electron density was observed between Pt2+ and the side chain Sδ of M782, with the Sδ replacing one of the Pt2+-coordinating Cl− ions, indicating the formation a Pt2+–S coordinate bond. Subsequent refinement cycles were performed by restraining the Pt2+–S, Pt2+–N, and Pt2+–Cl− bond lengths to averaged values seen in four-coordinated Pt(II) complexes and the four ligands to arrange in a square planar geometry (26). Because no coordinate bond was observed between etoplatin-N2α and protein, the stereochemical parameters derived from the high-resolution structure of cisplatin were applied to restrain the dichlorodiammineplatinum(II) moiety during the refinement of etoplatin-N2α-bound structure. Additional details on model building and refinement of the etoplatin-bound structures are described in the Supplementary Material and Methods. The data collection and refinement parameters are listed in Table 1.
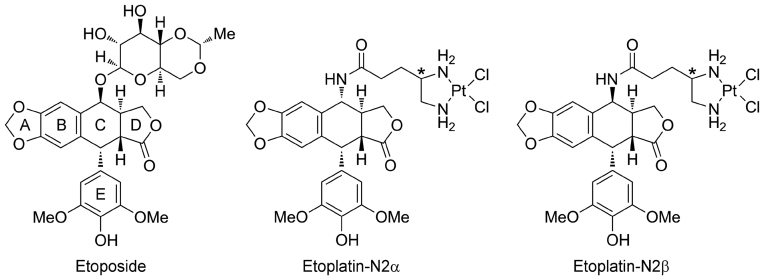
Figure 1.
Chemical structures of etoposide and the two etoplatins synthesized in this study. The polycyclic aglycone (rings A–D) and pendant ring (E-ring) of etoposide are labeled. A cis-dichlorodiammineplatinum(II) moiety was introduced via an amide linkage to the C4 position of the aglycone core in α and β configuration about the E-ring to produce etoplatin-N2α and N2β, respectively. Both etoplatins contain an additional chiral center (marked with asterisks) whose chirality was not specified during synthesis.
Table 1. Summary of crystallographic analysis.
| Protein | hTop2βcore | hTop2αcore | |
|---|---|---|---|
| PDB ID code | 5GWJ | 5GWI | 5GWK |
| Ligand | Etoplatin-N2β | Etoplatin-N2α | Etoposide |
| Space group | P 21 | P 21 2 21 | |
| Unit cell | |||
| Dimensions | |||
| a, b, c (Å), β (degrees) | 80.18, 177.0, 94.41, 111.5 | 79.79, 176.97, 94.53, 112.3 | 105.11, 126.16. 198.86 |
| Data collection | |||
| Resolution range (Å) | 29.64–2.57 (2.66–2.57) | 29.68–2.74 (2.84–2.74) | 27.42–3.15 (3.27–3.15) |
| Observed reflections | 290555 | 226170 | 309096 |
| Unique reflections | 77673 (7441) | 62535 (6108) | 45909 (4494) |
| Multiplicity | 3.7 (3.7) | 3.6 (3.4) | 6.7 (6.7) |
| Completeness (%) | 99.9 (99.8) | 98.2 (96.7) | 99.2 (99.7) |
| Mean I/sigma(I) | 15.7 (2.8) | 13.6 (3.0) | 24.6 (3.7) |
| Rsyma (last shell) (%) | 0.08 (0.484) | 0.087 (0.469) | 0.062 (0.48) |
| CC1/2 | 0.952 (0.801) | 0.957 (0.815) | 0.988 (0.947) |
| Data refinement | |||
| Wilson B-factor | 39.94 | 49.32 | 92.61 |
| Rcrysb (%) | 0.18 | 0.21 | 0.20 |
| Rfreeb (%) | 0.22 | 0.25 | 0.24 |
| RMSD (bonds) | 0.004 | 0.002 | 0.002 |
| RMSD (angles) | 0.743 | 0.526 | 0.566 |
| Ramachandranc | |||
| favored (%) | 96.5 | 96.5 | 96.1 |
| outliers (%) | 0.0 | 0.0 | 0.0 |
| Clashscore | 3.54 | 3.16 | 3.26 |
| Average B-factor | 46.6 | 51.7 | 108.1 |
Statistics for the highest-resolution shell are shown in parentheses.
aRsym = (Σ|Ihkl -〈I〉|)/(ΣIhkl), where the average intensity〈I〉is taken overall symmetry equivalent measurements and Ihkl is the measured intensity for any given reflection.
bRcryst = (Σ||Fo| - k|Fc||)/(Σ|Fo|). Rfree = Rcryst for a randomly selected subset (5%) of the data that were not used for minimization of the crystallographic residual.
cCategories were defined by PHENIX (24). All non-glycine residues are included for this analysis.
Structural modeling of etoplatin-stabilized hTop2α cleavage complexes
Structural models were constructed by first replacing the bound etoposide molecules in the crystal structure of hTOP2αcore-DNA-etoposide with etoplatin-N2β and N2α, respectively, under the assumption that the aglycone core and E-ring of the three compounds (Figure 1) share the same binding mode. The linker that connects the reactive dichlorodiammineplatinum(II) moiety to the aglycone C-ring was also assumed to adopt the conformation observed in the crystal structures of hTop2βcore–DNA–etoplatin–N2β and hTop2βcore–DNA–etoplatin–N2α complexes. Next, all energetically accessible rotamers of the hTOP2α-specific M762 were surveyed to identify side-chain conformations that allow its Sδ to overlay on and substitute for one of the Cl− ions of the dichlorodiammineplatinum(II) moiety as a ligand for Pt2+. The Pt2+–Sδ bond length was then restrained at ∼2.3 Å during subsequent energy minimization using the ‘In Situ Ligand Optimization’ and ‘Calculate Binding Energies’ modules of Discovery Studio 4.1 to idealize the drug-binding mode. For the additionally designed Pt2+ derivatives, structural modeling was conducted using the same procedure except that the conformations of the linker and the M762 side chain were both adjusted to allow the formation of Pt2+–Sδ bond.
Supercoil relaxation and DNA cleavage assays
The supercoil relaxation reaction (20 μl) was conducted at 37°C for 30 min in a buffer containing 10 mM Tris–HCl pH 8.0, 100 mM KCl, 0.1 mM EDTA, 5 mM MgCl2, 1 mM adenosinetriphosphate (ATP), 100 μg/ml bovine serum albumin (BSA) and 300 ng supercoiled (SC) pRYG plasmid DNA, with a titration of the indicated drug from 10 to 250 nM. A total of 120 ng/μl of hTop2△CTD was added to start the reaction. The reaction was terminated by the addition of 0.4 μl of 0.5% sodium dodecyl sulphate (SDS) and 0.8 μl of 250 mM EDTA and incubated at 37 °C for 1 h. The DNA cleavage reaction (20 μl) was conducted at 37 °C for 30 min in a buffer composed of 1 mM ATP, 100 μg/ml BSA, 250 ng of HindIII-linearized pRYG plasmid DNA, 20 nM of indicated drug and 1.2 μg/μl of hTop2△CTD. The cleavage reaction was terminated by the addition of 2 μl of 5% SDS followed by 2 μl of 250 mM EDTA. The enzyme was digested by the addition of 2 μl proteinase K (0.8 mg/ml) and incubation at 45 °C for 30 min. For reactions aiming to test the reversibility of DNA cleavage by the enzymes, EDTA was added to the reactions prior to adding SDS at the termination step. The resultant reaction mixtures from both assays were resolved by electrophoresis on 1% agarose gel followed by standard ethidium bromide (EtBr) staining analysis.
Measuring Pt2+ concentration by ICP-MS
To measure the release of etoplatin-N2α and -N2β from the Top2 cleavage complex, we employed inductively coupled plasma mass spectrometry (ICP-MS) (27), a superior technique for quantitative and ultrasensitive elemental analysis, to detect the platinum signal from the released drugs. The drug-stabilized Top2cc (in a 20 μl reaction) was prepared in a buffer containing 1 mM ATP, 100 μg/ml BSA, 1 μM of a 30 bp double-stranded DNA, 0.5 μM etoplatin-N2α or -N2β and 1.2 μM of hTop2α△CTD. After incubating the mixture at 37 °C for 30 min, guanidine hydrochloride (GdnHCl) was added to a final concentration of 1.2 M. Aliquots (10 μl) were sampled at different time points and then diluted with ddH2O to a volume of 100 μl. The sample was then filtered using a 10 kDa-cutoff filter membrane (UFC501096, Amicon Ultra 0.5 ml centrifugal filter) to separate the released (and thus membrane-permeable) drug from those that remained associated with Top2cc. A total of 20 μl of flow-through containing the released drugs was diluted again with ddH2O to a final volume of 3 ml. The resultant sample was then subjected to platinum content determination with an Agilent 7700× ICP-MS in the instrument center of National Taiwan University.
Cell lines, comet assay, quantitative measurement and statistical analysis
Parental HL-60 and its hTop2-deficient HL-60/MX2 leukemia cell lines were obtained from the American Type Culture Collection (ATCC) (28). Both cells were cultivated in Roswell Park Memorial Institute-1640 medium (RPMI) 1640 supplemented with 10% fetal calf serum. A total of 100 units/ml penicillin, 100 μg/ml streptomycin and 2 mM glutamine in a 5% CO2 incubator at 37°C. Comet assay was used to determine chromosome DNA breaks (29). Briefly, cells (2 × 105) were treated with etoposide or etoplatins for 1 h, then the cells were collected and re-suspended in 1 ml ice-cold 1× phosphate-buffered saline buffer. Drug-treated cells (50 μl) were mixed with 0.1 ml of pre-warmed low-melting-point agarose (0.7%) and loaded onto a fully frosted slide that had been pre-coated with 0.7% agarose. The slide was then covered with a coverslip and subsequently submerged in the pre-chilled lysis solution for 0.5 h at 4°C. After soaking with electrophoresis buffer for 40 min, the slides were subjected to electrophoresis for 20 min at 2 V/cm. DNA on the slides was stained with SybrGold (Molecular Probes, USA), visualized under a fluorescent microscope (Nikon Eclipse 80i, assay, over 100 cells were typically scored to calculate the percentage (%) of cell with comet tails. The quantitative results were analyzed and graphed using Prism 6.0 (GraphPad Software Inc.) and statistical analyses were performed with the unipolar paired student’s t-test. The data were considered statistically significant (P-values > 0.05, marked as * and 0.01, **) or very significant (P-values > 0.001, ***).
RESULTS
Structure-based design of an hTop2-targeting organoplatinum compound
The anticancer drug etoposide is an ideal candidate for testing the suitability of employing the Pt2+ coordinate chemistry in hTop2-targeting due to the well comprehended structure-activity relationships regarding its three constituting moieties (Figure 1) (16). The tetracyclic aglycone core composed of rings A–D mediates DNA intercalation and the appended E-ring provides specific interactions with protein residues located on the DNA minor groove side; both moieties are required for optimal drug action and are sensitive to modifications. Conversely, the pocket that houses the glycosidic group on the DNA major groove side not only is spacious enough to accommodate structurally distinct chemical groups, but also harbors potentially Pt2+-reactive methionine residue(s) (14,19). Replacing the glycosidic moiety with a Pt2+-containing group may thus allow for the formation of Pt2+–thioether bond between the drug and hTop2 isoforms. A diammine linker has previously been used successfully to introduce Pt2+ into podophyllotoxin (30). Our modeling analysis suggested that adjusting the length of the reported linker should place the Pt2+ within a favorable distance for conjugating to a nearby methionine and confers the resulting compounds potent Top2-poisoning activity. We propose to name these compounds etoplatins, representing Pt2+-conjugated etoposide derivatives. Two first generation etoplatins, N2β and N2α, which differ in chirality by which the diammine linker attaches to the C4 position of the aglycone core, were designed and synthesized (Figure 1 and Supplementary Schemes S1–3). The synthesis of etoplatin-N2β and N2α (5β and 5α, respectively) started with the preparation of the platinum(II)-diamino-carboxylic acid complex 2 followed by the amide bond formation with 4-amino-4-deoxy-4′-O-demethylpodophyllotoxin (4). The platinum(II)-4,5-diaminovaleric acid complex (PtCl2(N,N-Dav), 2) was obtained from the reaction of potassium tetrachloroplatinate(II) with 4,5-diaminovaleric acid 1 (31). The (4S)- and (4R)-4-amino-4-deoxy-4′-O-demethylpodophyllotoxins (4β and 4α, respectively) were prepared from 4′-O-demethyl-4-epi-podophyllotoxin 3 according to literature procedure (32). Subsequently, the amide bond formation between PtCl2(N,N-Dav) (2) and 4-amino-4-deoxy-4′-demethylpodophyllotoxins (4β and 4α, respectively) was accomplished by the reaction with N-ethyl-N'-(3-dimethylaminopropyl)carbodiimide hydrochloride and 1-hydroxybenzotriazole in N,N-dimethylformamide to produce etoplatin-N2β and N2α.
Etoplatin-N2β potently inhibits the supercoil relaxation activity of human Top2 isoforms
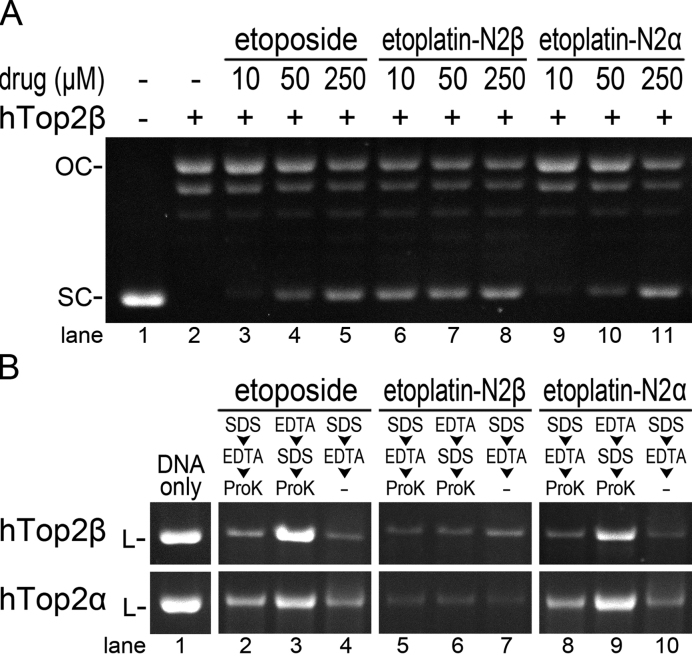
To examine the effects of etoplatins on the catalytic functions of Top2, we first compared the potency of these platinum organometallic compounds in blocking the Top2-mediated DNA relaxation to that of etoposide (Figure 2A and Supplementary Figure S1). While etoplatin-N2α and etoposide displayed similar inhibitory activities, a 25-fold lower concentration of etoplatin-N2β is sufficient to produce a comparable effect, indicating that etoplatin-N2β is significantly more effective in inhibiting the relaxation activity of both hTop2α and hTop2β. Given that etoplatin-N2β is produced by replacing the glycosidic moiety of etoposide with a thioether-directed, Pt2+-containing reactive center (Figure 1) and that both human Top2 isoforms exhibited increased sensitivity towards etoplatin-N2β, we reasoned that the Pt2+ center of etoplatin-N2β most likely forms a coordinate bond with the side-chain thioether moiety of Met766 in hTop2α and the spatially equivalent Met782 in hTop2β (14,19). In contrast, the N2α epimer appears to lack the capacity to form a coordinate bond with Top2 and like etoposide, it stabilizes the Top2cc mainly by non-covalent interactions. Together, these results suggest that the formation of a Pt2+–thioether coordinate bond is sensitive to the chirality by which the diammine linker attaches to the aglycone core of etoposide. The enhanced inhibition activity of etoplatin-N2β may be attributed to the existence of a coordinate bond in the resulting drug-arrested Top2cc.
Figure 2.
Etoplatin-N2β, but not the N2α epimer, more potently inhibits the supercoil relaxation activity of Top2 by inducing the formation of ethylenediaminetetraacetic acid (EDTA)-resistant DNA breaks. (A) Inhibition of the relaxation activity of hTop2 by etoposide and etoplatins. Each relaxation reaction contains 300 ng of supercoiled (SC) pRYG plasmid DNA. The enzyme-positive reactions contain 80 ng of hTop2β△CTD (designated as hTop2β). OC stands for open circle (full-relaxed product produced by Top2) DNA. (B) The DNA cleavage assay shows the production of EDTA-resistant, hTop2-mediated DNA breaks in the presence of etoplatin-N2β. Each cleavage reaction contains 250 ng of HindIII-linearized (L) pRYG plasmid DNA. A total of 1.2 μg of hTop2β△CTD (upper panel, designated as hTop2β) or hTop2α△CTD (lower panel, designated as hTop2α) was added to the respective enzyme-positive reactions. To stop the cleavage reaction, Sodium dodecyl sulphate (SDS) and EDTA were added in the indicated order and the denatured enzyme was removed by proteinase K digestion. Lanes labeled with -ProK indicates no proteinase K treatment after the reaction was stopped. The disappearance and reemergence of the linear substrate DNA (L) indicate the production and resealing of DNA breaks, respectively. Refer to Supplementary Figure S2 for the full-sized image of this gel.
The Top2-mediated DNA breaks arrested by etoplatin-N2β are practically irreversible
The Top2-mediated double-strand DNA breaks formed in the presence of clinically active hTop2-targeting anticancer drugs are inherently reversible; the two enzyme-tethered, complementary cohesive DNA ends harbored in a Top2cc can be rejoined under conditions where the reversal of transesterification reaction is favored (33–35). For example, exposing the drug-stabilized Top2cc to the Mg2+ chelator EDTA before the Top2 activity is permanently blocked by the sequential addition of protein denaturant and protease is known to promote DNA relegation and efficiently restores the integrity of the fragmented DNA, a common technique for assessing the reversibility of Top2cc (34,35). The speculation that a Pt2+–thioether coordinate bond is formed between etoplatin-N2β and Top2 predicts an enhanced stability of the resulting cleavage complex, presumably less reversible and thus more resistant to EDTA treatment. Indeed, while the DNA breaks induced by etoposide can be readily reversed by pre-treating the cleavage complex with EDTA, as indicated by the disappearance of the smeared DNA fragments and restoration of the full-length linear substrate DNA, the breakage resulted from etoplatin-N2β-mediated hTop2 poisoning cannot be resealed (Figure 2B and Supplementary Figure S2). Conversely, the N2α epimer appears to lack the capacity to form a coordinate bond with either human Top2 isoform and like etoposide, it stabilizes the Top2cc mainly by non-covalent interactions (lane 9 in Supplementary Figure S2 and Figure 2B). The production of irreversible, EDTA-resistant DNA breaks by etoplatin-N2β is consistent with its increased potency and provides additional support for its function as a coordinate bond-forming inhibitor.
Crystallographic analysis of hTop2β in complexes with DNA and etoplatins
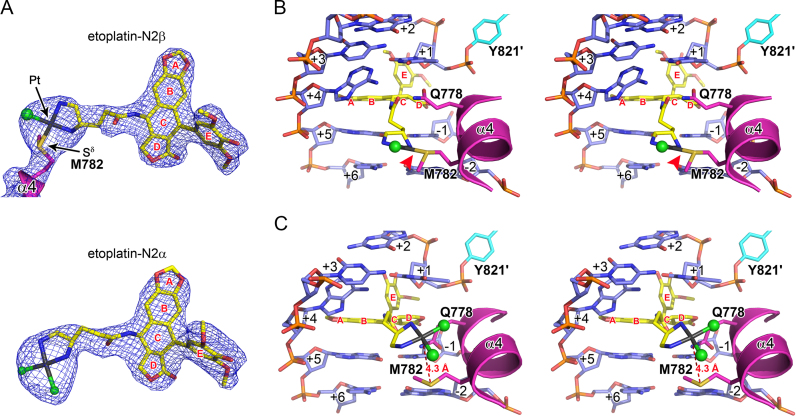
To confirm the proposed mechanisms of action for etoplatins, we performed X-ray crystallographic analysis on the etoplatin-stabilized cleavage complexes of hTop2β. Similar to etoposide (14), both etoplatins trap the Top2cc by targeting the enzyme-mediated DNA breaks, with the aglycone core intercalating between the base pairs flanking the cleavage site and the E-ring protruding toward the DNA minor groove to interact with surrounding residues (Figure 3B and C). As expected, the diamine linker extends towards the DNA major groove side and places the reactive dichloroplatinum(II) moiety in the general vicinity of the methionine residue(s) located in helix α4 of the Top2 winged-helix domain. For etoplatin-N2β, one of the chloride ions is replaced by the methionyl Sδ of M782 in hTop2β with clear electron density connecting Sδ and Pt2+, indicating that a coordinate bond with a refined bond length of 2.3 Å is formed (Figure 3A and Supplementary Figure S3A). Due to the structural constraints imposed by the alternative stereochemistry at the C4 chiral center, the dichloroplatinum(II) moiety of etoplatin-N2α is directed closer to the +1/+4 bp, which positions the Pt2+ more distantly (∼4.3 Å) from the methionyl Sδ (Figure 3C and Supplementary Figure S3B). No coordinate bond formation was observed and both of the Pt2+-ligating chloride ions are retained in etoplatin-N2α. Although the crystal structures of etoplatin-bound hTop2α are not yet available, structural modeling analysis predicted that hTop2α would form a coordinate bond with the bound etoplatin-N2β via the methionyl Sδ of M766 (corresponding to M782 in hTop2β) (Supplementary Figure S4A). In contrast, etoplatin-N2α would not coordinate with either M766 or M762 because their Sδ are too distant from the coordination sphere of Pt2+ to replace the Cl− ligand (Supplementary Figure S4B and C). Together, the results obtained from crystallographic and modeling analyses provide convincing evidence that etoplatin-N2β acts as a potent and irreversible poison of human Top2 isoforms through its coordinate bond forming capability.
Figure 3.
Both etoplatin-N2β and -N2α bind to the DNA cleavage sites in the hTop2βcc crystal structure as etoposide but only etoplatin-N2β forms the irreversible Pt2+–thioether coordinate bond. (A) The electron density maps of the bound drugs in the crystal structures of hTop2βcore-derived cleavage complexes stabilized by etoplatin-N2β (upper panel) and etoplatin-N2α (lower panel). The final 2DFo–mFc maps (at 1.5 σ) of the respective drugs are shown as blue meshes. Continuous electron density can be observed between Pt2+ and the Sδ of M782, indicating the formation of a coordinate bond. (B and C) Stereo views of the drug-binding site in the etoplatin-N2β- and etoplatin-N2α-stabilized cleavage complexes derived from hTop2βcore, respectively. The Pt2+–thioether coordination in the etoplatin-N2β-stabilized structure in B is specified by the red arrow. DNA is shown in sticks representation (blue) and labeled with positive and negative numbers to designate nucleotides downstream and upstream, respectively, of the scissile phosphate. The bound drugs are shown in sticks representation (yellow) with the Pt2+ in gray and Cl− in green spheres. The drug’s aromatic rings are labeled in red letters. The two protein chains are shown in a cartoon/stick representation and colored in magenta and cyan, respectively. The scissile phosphate-linked active site Y821 is labeled with a prime to specify that this residue is from the second protein chain.
We have shown previously that the conventional non-covalent Top2-targeting drugs can be released from the crystallized Top2 cleavage complex by an overnight soaking of the respective crystals in a drug-free substitute mother liquor and that the binding of a different drug can be achieved by another overnight soaking of these pre-treated crystals in a solution containing the new drug (16). When subjected to this two-step soaking procedure with another hTop2-targeting drug mitoxantrone being added in the second soaking solution, we observed the persistent presence of etoplatin-N2β in the DNA cleavage site even when the length of the second soaking step was extended to 1 week (Supplementary Figure S5). The bound N2α epimer, in contrast, behaved like etoposide and can be efficiently replaced following the same treatment. This result demonstrates that the coordinate bond formation confers remarkable stability to the etoplatin-N2β-stabilized Top2 cleavage complex, which in turn contributes to its enhanced Top2-poisoning activity.
The coordinate tether between etoplatin-N2β and hTop2 exhibits a protein conformation-dependent reversibility
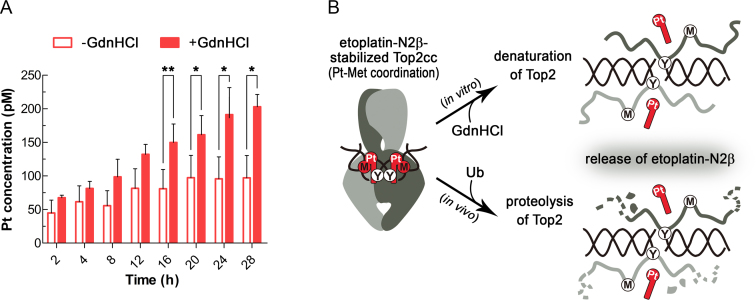
Earlier solution nuclear magnetic resonance analysis of the methionine-conjugated platinum(II) diammine complexes revealed that the mono-methionine derivative was susceptible to competition by guanosine 5'-monophosphate, as shown by the release of the Pt2+-ligated methionine, whereas the bis-methionine compound was substantially more resistant to ligand replacement and thus displayed greater stability in an aqueous solution (36). The presence of only a single Pt2+–thioether linkage between etoplatin-N2β and hTop2 suggests that the resulting tetracoordinate Pt2+ center is chemically similar to the more labile mono-methionine compound. Nevertheless, the coordinate bond formed between the cleavage site-bound etoplatin-N2β and hTop2βcore exhibited an outstanding stability (Supplementary Figure S5). We speculated that the compound’s podophyllotoxin moiety may contribute substantially to the enhanced stability by not only delivering the Pt2+ center to react with the thioether group of M782 in a site-specific manner, but also providing non-covalent interactions to sustain the Pt2+–thioether bond by keeping the metal/ligand pair in close proximity. To examine the validity of this hypothesis, we tested whether the bound etoposide-N2β can dissociate from Top2cc upon GdnHCl-induced protein denaturation (Figure 4A). As expected, an increase in the Pt2+ concentration from the detached etoplatin-N2β, as judged by its ability to pass through an ultrafiltration membrane (M.W. cutoff: 10 kDa), was observed in the presence of GdnHCl (Figure 4A). In contrast, for the untreated sample, no significant membrane permeable signal of Pt2+ could be detected after 28 h of incubation at 37°C. This result supports the importance of non-covalent interactions in stabilizing the coordinate bond. Moreover, it can be inferred that the strength of a metal-protein linkage may be reduced upon disruption of the protein structure. Therefore, the apparently irreversible trapping of Top2cc by etoplatine-N2β exhibits protein conformation-dependent reversibility.
Figure 4.
The etoplatin-N2β-mediated Pt2+–thioether coordination relies on the integrity of the hTop2cc structure. (A) The release of etoplatin-N2β (as reflected by the amount of membrane permeable Pt2+) from structurally intact guanidine hydrochloride (-GdnHCl) and denatured (+GdnHCl) Top2cc derived from hTop2α△CTD was quantified by inductively coupled plasma mass spectrometry (ICP-MS). The membrane permeable Pt2+ level of the +GdnHCl group gradually increases with time and was significantly higher than the -GdnHCl group started from 16 h. (*: P < 0.05, **: P < 0.01) (B) Cartoon representations illustrating that the release of etoplatin-N2β from Top2cc upon GdnHCl treatment (in vitro route) may be achieved by targeting Top2 to the 26S proteasome for degradation (in vivo route). Specifically, the trapped Top2cc on genomic DNA in the cell will be ubiquitinated (Ub) and channeled to the 26S proteasome for degradation (46,47). Upon structural disruption of its binding site, etoplatin-N2β can no longer form stable coordination with the targeting methionine, allowing its release from the protein.
Cellular Top2s can be targeted by etoplatin-N2β but at a reduced potency
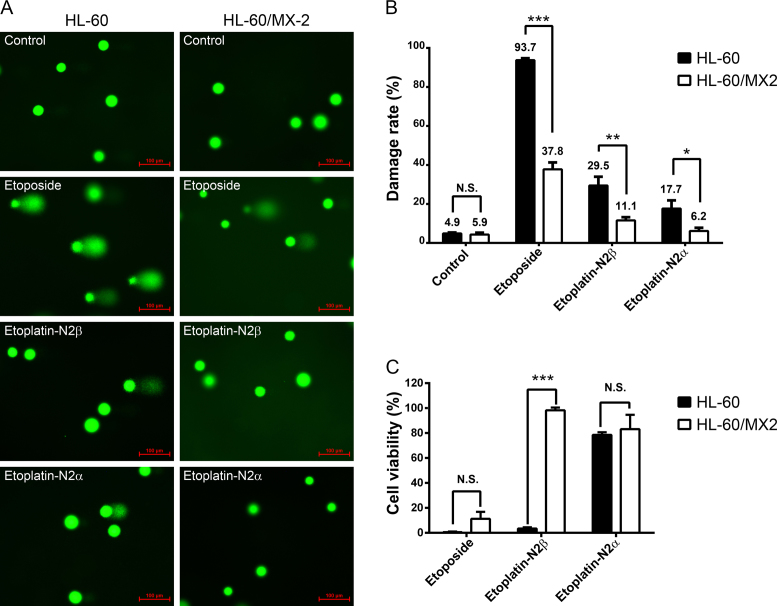
The enhanced potency of etoplatin-N2β in poisoning human Top2s, as observed in the various in vitro assays shown above (Figure 2; Supplementary Figures S1 and 2), implicated that this new compound might exhibit a stronger cytotoxicity than etoposide. To determine whether hTop2α and hTop2β can indeed be targeted by etoplatins in vivo, the comet and 4-day 3-(4,5-Dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT) cytotoxicity assays were performed using the human HL-60 and its hTop2-deficient HL-60/MX2 leukemia cell lines. It has been well documented that the mRNA and protein levels of both hTop2α and hTop2β are greatly reduced in HL-60/MX2 cells, thus rendering them more resistant to various hTop2-targeting drugs, including etoposide (28). As expected, etoplatin-N2β was found to be more effective than the N2α epimer in causing chromosomal DNA fragmentation and cancer cell killing (Figure 5 and Supplementary Table S1). In addition, the population of HL-60/MX2 cells with DNA damage was significantly lower than HL-60 cells after treatments with both etoplatin-N2β and N2α, which is consistent with the involvement of hTop2cc in causing the drug-induced DNA fragmentation. The finding that etoplatin-N2β caused >90% (98.3 ± 2.2) and <10% (3.5 ± 1.0) of cell death in HL-60 and HL-60/MX-2 cells, respectively, further suggests the drug-induced chromosomal breakage is a key determinant for the cytotoxicity of etoplatins (Figure 5C). However, these assays also revealed that etoposide was more potent than both etoplatins in inducing DNA damage and cell death (Figure 5 and Supplementary Table S1). We envision four possibilities that may account for the discrepancy between the results obtained from in vitro and cell-based assays. First, replacement of the glycosidic group by an amide-linked cis-dichlorodiammineplatinum(II) moiety may cause etoplatins to be less efficiently taken up or more avidly effluxed by cells than etoposide. Second, the reactive Pt2+ center is attached to the etoposide’s aglycone core via an amide linkage. It is possible that this amide bond may be hydrolyzed by an unknown amidase in cells, which would compromise the efficacy of the new compounds. Third, the efficacy of the new compounds may be suppressed by their interactions with other cellular components. For example, etoplatins may be modified and even deactivated by reacting with the thiol group of cellular glutathione. Finally, it is possible that only a small percentage of the new compounds enters the nucleus to interact with Top2, whereas the majority of them were trapped at other cellular compartments. Additional efforts would be required to pin down the actual cause(s) of this discrepancy.
Figure 5.
Like etoposide, etoplatins induced chromosomal DNA breaks and cancer cell death in a hTop2-dependent manner. (A) Levels of etoposide- and etoplatin-induced chromosomal DNA breaks are significantly reduced in the hTop2-deficient HL-60/MX2 cells compared to the parental HL-60 cells. It has been shown that HL-60/MX2, a mitoxantrone-resistant variant of HL-60 cell line, expresses much lower levels of both hTop2α and hTop2β (28). Cells were treated with 50 μM of etoposide or etoplatins for 1 h, comet assay were then carried out to measure chromosomal DNA breaks and quantified data were shown in (B). In the microscopic images, each green circular head or comet (head + tail) image represents one cell. The green dots are indicative of intact nucleoids containing SC loops of chromosomal DNA attached to the nuclear matrix. Upon drug treatments, damaged nuclear DNA becomes relaxed due to the presence of DNA breaks resulting from drug-stabilized Top2cc and a tail of damaged DNA migrates toward the anode during electrophoresis, leading to the comet-like appearance. Values as determined by the percentage of cells with DNA damage are presented as mean ± SD (n = 3). (C) Etoplatin-N2β exhibits clear hTop2-dependent cancer cell killing activity. HL-60 leukemia cells were treated with 100 μM of etoposide, etoplatin-N2β or etoplatin-N2α, and after 4 days of incubation, MTT assay were performed to determine cytotoxicity. N.S. (not significant, P > 0.05); *P < 0.05; **P < 0.01; ***P < 0.001.
DISCUSSION
In this work, we conducted structure-based modification of the clinically active anticancer drug etoposide to produce an organoplatinum compound (Figure 1), etoplatin-N2β, which exhibits potent Top2-poisoning activity (Figure 2). The podophyllotoxin core of this new compound offers binding specificity with the Top2-mediated DNA cleavage site and the appended cis-dichlorodiammineplatinum(II) moiety strengthens the drug–protein interaction by forming a Pt2+–thioether coordinate bond with a flanking methionine residue (Figure 3A and B). Our results clearly show that selective targeting of a specific methionine residue in the protein can be achieved by exploiting the Pt2+ coordination chemistry and that the combination of a single Pt2+–thioether linkage and non-covalent interactions between the drug and protein is sufficient to increase the stability of the resulting Top2–DNA–drug ternary complex.
Patients receiving Top2-targeting anticancer drugs are at risk of developing therapy-related secondary leukemia and cardiac problems (37,38). Mounting evidence suggests that the undifferentiated targeting of both human Top2 isoforms by these drugs is likely the main cause of adverse effects: while poisoning of hTop2α is sufficient for killing cancer cells, the induction of hTop2β-mediated DNA breaks and chromosome translocation events may result in deleterious side effects (39–42). To overcome these problems, it would be clinically desirable to develop a hTop2α-specific drug without simultaneous targeting of hTop2β. Structural analysis of the etoposide-bound hTop2α cleavage complex (Table 1) revealed that, compared to the Pt2+-coordinating methionine that was either observed (M782 of hTop2β) or predicted (M766 of hTop2α) in this study, the thioether side chain of the hTop2α-specific methionine (M762) is located significantly closer to ring C of the bound podophyllotoxin core (Supplementary Figure S6). Therefore, we envision a shorter diammine linker with properly designed stereochemistry would deliver the reactive Pt2+ center towards M762 to achieve selective targeting of hTop2α over the β-isoform, which instead, possesses a glutamine (Q778) at the corresponding position. We have taken the first step toward this end by designing a series of synthesizable compounds with altered linker structure and length (Supplementary Figure S7). For all these compounds, structural modeling showed that the Pt2+ can be placed at ∼2.3 Å from the Sδ of M762 to allow coordination bond formation, whereas the simulated distances between Pt2+ and the Sδ of M766 are much longer (>5.0 Å) and thus preclude bond formation between the two. This analysis suggests that selective targeting of hTop2α via the isoform-specific M762 may be an achievable goal.
Top2-targeting drugs from different chemical classes may produce distinct DNA cleavage patterns, which may be rationalized in part by the presence of sequence-specific drug–DNA interactions (43). For example, the polar and van der Waals interactions formed between the glycosidic group of etoposide and the +5/−1 bp at the DNA cleavage site may explain the preference for having a GC base pair at this position (14). Since the glycosidic group was replaced by Pt2+ reactive center in etoplatins, it is reasonable to anticipate that each compound may produce its own unique Top2-mediated DNA cleavage patterns. Indeed, notable differences were recognized between the DNA cleavage patterns induced respectively by etoposide, etoplatin-N2β and N2α (Supplementary Figure S2). Structural analysis of the Top2cc stabilized by etoplatin-N2β revealed that the amide moiety of the diammine linker forms hydrogen bonds with both the +5/−1 and +4/+1 bp (Supplementary Figure S3A). In contrast, the diammine linker of etoplatin-N2α pushes against the +4/+1 bp, resulting in significant displacements of the +5/−1 and +4/+1 bp from their positions seen in the etoposide-bound structure (Supplementary Figure S3B). Given that the sequence of the substrate DNA used in our structural analyses was optimized for etoposide-induced cleavage, the aforementioned differences in drug–DNA interactions may implicate altered sequence preferences in DNA cleavage.
Unlike clerocidin, an archetypal Top2 inhibitor that irreversibly traps Top2cc by forming permanent covalent bonds with DNA (44,45), a potential advantage of employing Pt2+–thioether coordination chemistry in drug design is that the stability of the resulting coordinate tether formed between the drug and its target methionine appears to rely on the existence of drug-mediated non-covalent interactions (Figure 4A). Damage to the structural integrity of the drug binding pocket by protein denaturant is expected to abolish the non-covalent part of the interactions between drug and Top2cc. The accelerated release of etoplatin-N2β from Top2cc in the presence of GdnHCl indicates that the Pt2+–thioether linkage was weakened upon protein unfolding (Figure 4B, top panel). Although in this experiment the dissociation of Top2cc-attached etoplatin-N2β is initiated rather artificially by the addition of GdnHCl, we speculate a similar protein unfolding-triggered drug dissociation event may also take place in vivo. The repair of double-strand DNA breaks harbored by the drug-arrested Top2cc involves unfolding and 26S proteasome-dependent proteolytic removal of Top2 (46,47), which would disrupt the non-covalent drug-protein contacts and renders the Pt2+–thioether bond reversible (Figure 4B, bottom panel). Unlike covalent inhibitors that usually form irreversible adducts with proteins or nucleic acids, the organoplatinum compounds display a protein conformation-dependent reversibility, and hence may be less toxic and antigenic.
Taken together, our results suggest a general approach for developing organoplatinum compounds that permit targeting of a specific methionine residue in proteins: a successful compound should have a collection of suitable pharmacophores that are recognized by the target protein via the formation of a non-covalent complex. The dichloroplatinum(II) moiety of the compound would then coordinate with a nearby methionine side chain to further stabilize the drug–protein interaction. This strategy is expected to allow for effective targeting of any protein with a methionine-containing drug-binding pocket. In addition to the successful development of etoplatin-N2β as a potent hTop2 poison, it appears that bacterial DNA gyrase and a drug-resistant epidermal growth factor receptor (EGFR) variant carrying a T790M mutation would also be ideal candidates for illustrating the applicability of using Pt2+–thioether coordination in drug design. The binding site of Novel Bacterial Type II Topoisomerase Inhibitors (NBTIs) that target bacterial DNA gyrase features the presence of several methionine residues (48); introducing a Pt2+-containing group into NBTIs may further strengthen protein–drug interaction. Similarly, the T790M mutation, located in the EGFR’s ATP-binding site, weakens the interaction between EGFR and the tyrosine kinase inhibitor gefitinib (49). A Pt2+-derivatized gefitinib with the ability to coordinate with M790 may restore its association with EGFR. In summary, the integration of the dichloroplatinum(II) moiety into an existing drug, as shown in the development of etoplatin-N2β, may represent an effective technical shortcut for boosting the drug’s bioactivity.
ACCESSION NUMBERS
Atomic coordinates and structure factors have been deposited in the PDB with accession codes 5GWJ (human topoisomerase IIβ with DNA and etoplatin-N2β), 5GWI (human topoisomerase IIβ with DNA and etoplatin-N2α) and 5GWK (human topoisomerase IIα with DNA and etoposide).
Supplementary Material
ACKNOWLEDGEMENTS
We thank Prof Chun-Hua Hsu and our lab members for their technical support and commenting on the manuscript. Portions of this research were carried out at beamlines 15A1 and 13B1 of the National Synchrotron Radiation Research Center (Taiwan) and beamline SP12B2 of the SPring-8 (Japan). We are grateful to the staffs of Technology Commons in College of Life Science and Center for Systems Biology, National Taiwan University.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Ministry of Science and Technology [NSC101-2911-I-002-303, 103-2113-M-002-010-MY3, 104-2911-I-002-302, 106-2113-M-002-021-MY3]; National Research Program for Biopharmaceuticals [NSC101-2325-B-002-049]; National Taiwan University [104R7614-3, 104R7560-4]; Ministry of Education, Taiwan ROC, under the ATU plan (National Chung Hsing University) (to N.L.C.). Funding for open access charge: Ministry of Science and Technology, Taiwan.
Conflict of interest statement. None declared.
REFERENCES
- 1. Zarzycka B., Kuenemann M.A., Miteva M.A., Nicolaes G.A., Vriend G., Sperandio O.. Stabilization of protein-protein interaction complexes through small molecules. Drug Discov. Today. 2016; 21:48–57. [DOI] [PubMed] [Google Scholar]
- 2. Potashman M.H., Duggan M.E.. Covalent modifiers: an orthogonal approach to drug design. J. Med. Chem. 2009; 52:1231–1246. [DOI] [PubMed] [Google Scholar]
- 3. Singh J., Petter R.C., Baillie T.A., Whitty A.. The resurgence of covalent drugs. Nat. Rev. Drug Discov. 2011; 10:307–317. [DOI] [PubMed] [Google Scholar]
- 4. Lavergne S.N., Park B.K., Naisbitt D.J.. The roles of drug metabolism in the pathogenesis of T-cell-mediated drug hypersensitivity. Curr. Opin. Allergy Clin. Immunol. 2008; 8:299–307. [DOI] [PubMed] [Google Scholar]
- 5. Uetrecht J. Immune-mediated adverse drug reactions. Chem. Res. Toxicol. 2009; 22:24–34. [DOI] [PubMed] [Google Scholar]
- 6. Johnson D.S., Weerapana E., Cravatt B.F.. Strategies for discovering and derisking covalent, irreversible enzyme inhibitors. Future Med. Chem. 2010; 2:949–964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Kettle S.F.A. Physical Inorganic Chemistry: a Coordination Chemistry Approach. 1996; Heidelberg, Berlin: Springer-Verlag Berlin. [Google Scholar]
- 8. Vos S.M., Tretter E.M., Schmidt B.H., Berger J.M.. All tangled up: how cells direct, manage and exploit topoisomerase function. Nat. Rev. Mol. Cell Biol. 2011; 12:827–841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Chen S.H., Chan N.L., Hsieh T.S.. New mechanistic and functional insights into DNA topoisomerases. Annu. Rev. Biochem. 2013; 82:139–170. [DOI] [PubMed] [Google Scholar]
- 10. Deweese J.E., Osheroff N.. The DNA cleavage reaction of topoisomerase II: wolf in sheep's clothing. Nucleic Acids Res. 2009; 37:738–748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Nitiss J.L. Targeting DNA topoisomerase II in cancer chemotherapy. Nat. Rev. Cancer. 2009; 9:338–350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Pommier Y., Marchand C.. Interfacial inhibitors: targeting macromolecular complexes. Nat. Rev. Drug Discov. 2012; 11:25–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Pommier Y. Drugging topoisomerases: lessons and challenges. ACS Chem. Biol. 2013; 8:82–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Wu C.C., Li T.K., Farh L., Lin L.Y., Lin T.S., Yu Y.J., Yen T.J., Chiang C.W., Chan N.L.. Structural basis of type II topoisomerase inhibition by the anticancer drug etoposide. Science. 2011; 333:459–462. [DOI] [PubMed] [Google Scholar]
- 15. Miles T.J., Hennessy A.J., Bax B., Brooks G., Brown B.S., Brown P., Cailleau N., Chen D., Dabbs S., Davies D.T. et al. . Novel hydroxyl tricyclics (e.g., GSK966587) as potent inhibitors of bacterial type IIA topoisomerases. Bioorg. Med. Chem. Lett. 2013; 23:5437–5441. [DOI] [PubMed] [Google Scholar]
- 16. Wu C.C., Li Y.C., Wang Y.R., Li T.K., Chan N.L.. On the structural basis and design guidelines for type II topoisomerase-targeting anticancer drugs. Nucleic Acids Res. 2013; 41:10630–10640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Blower T.R., Williamson B.H., Kerns R.J., Berger J.M.. Crystal structure and stability of gyrase-fluoroquinolone cleaved complexes from Mycobacterium tuberculosis. Proc. Natl. Acad. Sci. U.S.A. 2016; 113:1706–1713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Veselkov D.A., Laponogov I., Pan X.S., Selvarajah J., Skamrova G.B., Branstrom A., Narasimhan J., Prasad J.V., Fisher L.M., Sanderson M.R.. Structure of a quinolone-stabilized cleavage complex of topoisomerase IV from Klebsiella pneumoniae and comparison with a related Streptococcus pneumoniae complex. Acta Crystallogr. D Struct. Biol. 2016; 72:488–496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Wendorff T.J., Schmidt B.H., Heslop P., Austin C.A., Berger J.M.. The structure of DNA-bound human topoisomerase II alpha: conformational mechanisms for coordinating inter-subunit interactions with DNA cleavage. J. Mol. Biol. 2012; 424:109–124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Wasserman R.A., Austin C.A., Fisher L.M., Wang J.C.. Use of yeast in the study of anticancer drugs targeting DNA topoisomerases: expression of a functional recombinant human DNA topoisomerase II alpha in yeast. Cancer Res. 1993; 53:3591–3596. [PubMed] [Google Scholar]
- 21. Austin C.A., Marsh K.L., Wasserman R.A., Willmore E., Sayer P.J., Wang J.C., Fisher L.M.. Expression, domain structure, and enzymatic properties of an active recombinant human DNA topoisomerase II beta. J. Biol. Chem. 1995; 270:15739–15746. [DOI] [PubMed] [Google Scholar]
- 22. Chen Y.T., Collins T.R., Guan Z., Chen V.B., Hsieh T.S.. Probing conformational changes in human DNA topoisomerase IIalpha by pulsed alkylation mass spectrometry. J. Biol. Chem. 2012; 287:25660–25668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Otwinowski Z., Minor W. et al. . Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 1997; 276:307–326. [DOI] [PubMed] [Google Scholar]
- 24. Adams P.D., Afonine P.V., Bunkoczi G., Chen V.B., Davis I.W., Echols N., Headd J.J., Hung L.W., Kapral G.J., Grosse-Kunstleve R.W. et al. . PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D Biol. Crystallogr. 2010; 66:213–221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Emsley P., Lohkamp B., Scott W.G., Cowtan K.. Features and development of Coot. Acta Crystallogr. D Biol. Crystallogr. 2010; 66:486–501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Melnik M., Mikus P.. Structure aspects of monomeric platinum coordination complexes. Mater. Sci. Appl. 2014; 5:512–547. [Google Scholar]
- 27. Ghezzi A., Aceto M., Cassino C., Gabano E., Osella D.. Uptake of antitumor platinum(II)-complexes by cancer cells, assayed by inductively coupled plasma mass spectrometry (ICP-MS). J. Inorg. Biochem. 2004; 98:73–78. [DOI] [PubMed] [Google Scholar]
- 28. Harker W.G., Slade D.L., Parr R.L., Holguin M.H.. Selective use of an alternative stop codon and polyadenylation signal within intron sequences leads to a truncated topoisomerase II alpha messenger RNA and protein in human HL-60 leukemia cells selected for resistance to mitoxantrone. Cancer Res. 1995; 55:4962–4971. [PubMed] [Google Scholar]
- 29. Lee C.H., Hsieh M.Y., Hsin L.W., Chen H.C., Lo S.C., Fan J.R., Chen W.R., Chen H.W., Chan N.L., Li T.K.. Anthracenedione-methionine conjugates are novel topoisomerase II-targeting anticancer agents with favorable drug resistance profiles. Biochem. Pharmacol. 2012; 83:1208–1216. [DOI] [PubMed] [Google Scholar]
- 30. Liu X., Zhang L.L., Xu X.H., Hui L., Zhang J.B., Chen S.W.. Synthesis and anticancer activity of dichloroplatinum(II) complexes of podophyllotoxin. Bioorg. Med. Chem. Lett. 2013; 23:3780–3784. [DOI] [PubMed] [Google Scholar]
- 31. Altman J., Wilchek M., Warshawsky A.. Platinum(II) complexes with 2,4-diaminobutyric acid, ornithine, lysine and 4,5-diaminovaleric acid. Inorg. Chim. Acta. 1985; 107:165–168. [Google Scholar]
- 32. Hansen H.F., Jensen R.B., Willumsen A.M., Norskovlauritsen N., Ebbesen P., Nielsen P.E., Buchardt O.. New compounds related to podophyllotoxin and congeners: synthesis, structure elucidation and biological testing. Acta Chem. Scand. 1993; 47:1190–1200. [DOI] [PubMed] [Google Scholar]
- 33. Liu L.F., Rowe T.C., Yang L., Tewey K.M., Chen G.L.. Cleavage of DNA by mammalian DNA topoisomerase II. J. Biol. Chem. 1983; 258:15365–15370. [PubMed] [Google Scholar]
- 34. Sander M., Hsieh T.. Double strand DNA cleavage by type II DNA topoisomerase from Drosophila melanogaster. J. Biol. Chem. 1983; 258:8421–8428. [PubMed] [Google Scholar]
- 35. Vann K.R., Sedgeman C.A., Gopas J., Golan-Goldhirsh A., Osheroff N.. Effects of olive metabolites on DNA cleavage mediated by Human Type II topoisomerases. Biochemistry. 2015; 54:4531–4541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Barnham K.J., Guo Z., Sadler P.J.. Stabilization of monofunctional platinum-nucleotide adducts: reactions of N-acetyl-L-methionine complexes with guanosine 5′-monophosphate and guanylyl(3′-5′)guanosine. J. Chem. Soc., Dalton Trans. 1996; 2867–2876. [Google Scholar]
- 37. Cowell I.G., Austin C.A.. Mechanism of generation of therapy related leukemia in response to anti-topoisomerase II agents. Int. J. Environ. Res. Public Health. 2012; 9:2075–2091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Rashidi A., Fisher S.I.. Therapy-related acute promyelocytic leukemia: a systematic review. Med. Oncol. 2013; 30:625. [DOI] [PubMed] [Google Scholar]
- 39. Errington F., Willmore E., Tilby M.J., Li L., Li G., Li W., Baguley B.C., Austin C.A.. Murine transgenic cells lacking DNA topoisomerase IIbeta are resistant to acridines and mitoxantrone: analysis of cytotoxicity and cleavable complex formation. Mol. Pharmacol. 1999; 56:1309–1316. [DOI] [PubMed] [Google Scholar]
- 40. Lopez-Lazaro M., Willmore E., Austin C.A.. Cells lacking DNA topoisomerase II beta are resistant to genistein. J. Nat. Prod. 2007; 70:763–767. [DOI] [PubMed] [Google Scholar]
- 41. Pommier Y., Leo E., Zhang H., Marchand C.. DNA topoisomerases and their poisoning by anticancer and antibacterial drugs. Chem. Biol. 2010; 17:421–433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Zhang S., Liu X., Bawa-Khalfe T., Lu L.S., Lyu Y.L., Liu L.F., Yeh E.T.. Identification of the molecular basis of doxorubicin-induced cardiotoxicity. Nat. Med. 2012; 18:1639–1642. [DOI] [PubMed] [Google Scholar]
- 43. Laponogov I., Sohi M.K., Veselkov D.A., Pan X.S., Sawhney R., Thompson A.W., McAuley K.E., Fisher L.M., Sanderson M.R.. Structural insight into the quinolone-DNA cleavage complex of type IIA topoisomerases. Nat. Struct. Mol. Biol. 2009; 16:667–669. [DOI] [PubMed] [Google Scholar]
- 44. Binaschi M., Zagotto G., Palumbo M., Zunino F., Farinosi R., Capranico G.. Irreversible and reversible topoisomerase II DNA cleavage stimulated by clerocidin: sequence specificity and structural drug determinated. Cancer Res. 1997; 57:1710–1716. [PubMed] [Google Scholar]
- 45. Pan X.S., Dias M., Palumbo M., Fisher L.M.. Clerocidin selectively modifies the gyrase-DNA gate to induce irreversible and reversible DNA damage. Nucleic Acids Res. 2008; 36:5516–5529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Xiao H., Mao Y., Desai S.D., Zhou N., Ting C.Y., Hwang J., Liu L.F.. The topoisomerase IIbeta circular clamp arrests transcription and signals a 26S proteasome pathway. Proc. Natl. Acad. Sci. U.S.A. 2003; 100:3239–3244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Azarova A.M., Lyu Y.L., Lin C.P., Tsai Y.C., Lau J.Y., Wang J.C., Liu L.F.. Roles of DNA topoisomerase II isozymes in chemotherapy and secondary malignancies. Proc. Natl. Acad. Sci. U.S.A. 2007; 104:11014–11019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Miles T.J., Hennessy A.J., Bax B., Brooks G., Brown B.S., Brown P., Cailleau N., Chen D., Dabbs S., Davies D.T. et al. . Novel tricyclics (e.g., GSK945237) as potent inhibitors of bacterial type IIA topoisomerases. Bioorg. Med. Chem. Lett. 2016; 26:2464–2469. [DOI] [PubMed] [Google Scholar]
- 49. Yun C.H., Boggon T.J., Li Y., Woo M.S., Greulich H., Meyerson M., Eck M.J.. Structures of lung cancer-derived EGFR mutants and inhibitor complexes: mechanism of activation and insights into differential inhibitor sensitivity. Cancer cell. 2007; 11:217–227. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.