Abstract
The Fabaceae family is considered as a model system for understanding chloroplast genome evolution due to the presence of extensive structural rearrangements, gene losses and localized hypermutable regions. Here, we provide sequences of four chloroplast genomes from the Lupinus genus, belonging to the underinvestigated Genistoid clade. Notably, we found in Lupinus species the functional loss of the essential rps16 gene, which was most likely replaced by the nuclear rps16 gene that encodes chloroplast and mitochondrion targeted RPS16 proteins. To study the evolutionary fate of the rps16 gene, we explored all available plant chloroplast, mitochondrial and nuclear genomes. Whereas no plant mitochondrial genomes carry an rps16 gene, many plants still have a functional nuclear and chloroplast rps16 gene. Ka/Ks ratios revealed that both chloroplast and nuclear rps16 copies were under purifying selection. However, due to the dual targeting of the nuclear rps16 gene product and the absence of a mitochondrial copy, the chloroplast gene may be lost. We also performed comparative analyses of lupine plastomes (SNPs, indels and repeat elements), identified the most variable regions and examined their phylogenetic utility. The markers identified here will help to reveal the evolutionary history of lupines, Genistoids and closely related clades.
Keywords: chloroplast genomes, Lupinus, functional gene relocation, repeated sequences, phylogeny
1. Introduction
The Fabaceae (or Leguminosae) is one of the largest flowering plant families, with ca. 19,500 herbaceous to tree species (ca. 751 genera) distributed in very diverse ecogeographical areas around the World.1–3 Because of their ability to establish specific associations with nitrogen-fixing rhizobial bacteria,4,5 many legume species are of great ecological and economic interest. They provide valuable biological nitrogen for better productivity and ecosystem functioning, and supply significant sources of protein for human and animal nutrition and health.6 Within Fabaceae, the Papillionoideae clade includes several major crops for human and animal consumption, such as soybean (Glycine), barrel medic (Medicago), bean (Phaseolus), cowpea (Vigna), chickpea (Cicer), pea (Pisum), peanut (Arachis), pigeon pea (Cajanus) and lupine (Lupinus). Increasing our knowledge of the evolutionary history of this family, as well as of the mechanisms involved in its physiological and ecological properties will improve management of natural and agricultural ecosystems and guide plant breeding programs.
During the last decade, our understanding of the structural and functional evolutionary dynamics of legume genomes increased significantly due to progress in Next Generation Sequencing (NGS) technologies. This recent sequencing of many plant plastomes revealed the unusual evolution of the Fabaceae, Geraniaceae and Campanulaceae plastomes.7–13 To date, 34 complete Fabaceae plastomes have been sequenced (including 17 in the last three years), mainly from Papilionoid lineages (25), and a few from the Cesalpinioid (5) and Mimosoid (4) lineages.14–18 Comparative analyses of Fabaceae plastomes showed that they have undergone major structural evolution compared with other plant families, including the lack of one inverted repeat (IR),19 a 51-kb inversion shared by most Papilionoid clades (including species from the IR lacking clade also called IRLC),20–22 a 78-kb inversion in Phaseolae,23–25 a 5.6-kb inversion in Milletia,26 a 36-kb inversion in the Genistoid clade15 and a 39-kb inversion in Robinia.16
Although gene content is relatively well conserved in angiosperm plastomes,12,27 it has been shown that several genes, such as accD,7psaI,12ycf4, rpl33,12clpP14,18 or rps1628, have been functionally lost in various legume lineages. Some of these chloroplast genes (accD and rps16), which are essential for plant survival,29–31 were shown to be functionally replaced by a nuclear gene.12,28
In contrast to the plastomes of most angiosperm families, Fabaceae plastomes have regions with accelerated mutation rates, including genic regions such as the rps16-ycf4 region in the IRLC clade12 or the clpP gene in Mimosoids.14,18 It has been suggested that this remarkable pattern of variation most likely results from the functional alteration of genes involved in DNA replication, repair and recombination,12,14,32 which may also facilitate the expansion of repeat sequences and the formation of structural rearrangements. For instance, the extensive reorganization of the plastid genome in Trifolium was correlated with an increase in repeat number,7 and the increase in size of the plastid genome in Mimosoids was correlated with tandem repeat expansions.14
Until recently, most of the knowledge of legume plastome evolutionary dynamics derived from model and crop plants in the Papilionoid lineage, and specifically the non-protein amino acid-accumulating (NPAA) clade (including Millettioids, Robinioids and IRLC).33 In the last three years, plastomes of other Papilionoids, Mimosoids and Cesalpinioids lineages have been sequenced and have provided additional insights into the unusual plastome evolution of the Fabaceae.14–18 In a few genera (Glycine, Lathyrus, Trifolium), the plastomes of several species were sequenced,12,17,34 contributing to a better understanding of the origin of specific structural variations. Localized hypermutations, gene losses and plastome size variations were identified as well as useful sequence resources were found for phylogenetic inference. However, additional sequencing efforts in key genera of the highly diverse legume family is essential for understanding key features of plastome evolution, and to resolve phylogenetic relationships at these taxonomic levels.
The Genistoid clade contains ∼18% of the 13,800 Papilionoid taxa.35 Within this poorly-studied lineage, the diverse Lupinus genus is considered as a good model system. Lupinus is composed of hundreds of annual and perennial herbaceous species and a few soft-woody shrubs and trees, which occur in a wide range of ecogeographical conditions. Lupines are mainly distributed in the New World (NW) (from Alaska to southern Chile and Argentina), whereas ∼20 species and subspecies are native to the Old World (OW) where two groups are distinguished: the smooth-seeded (circum-Mediterranean) and the rough-seeded lupines (scattered in North-equatorial Africa).36 In addition, this genus includes some crop species37 (Lupinus albus, Lupinus luteus, Lupinus angustifolius, Lupinus mutabilis), which are of growing interest due to their high seed protein content, their potential as nitrogen producers and for their health benefits.37,38 Molecular phylogenetic investigations using nuclear (ITS, nrDNA, LEGCYC1A, SymRK) and chloroplastic (rbcL, matK, trnL-trnF, trnL-trnT, trnL-intron, trnS-trnG) regions drastically improved knowledge of the evolutionary history of this complex genus.39–44 Many clades have been well circumscribed, and patterns of diversification were identified in both the NW and the OW. In spite of these significant advances, there are still uncertainties and unresolved relationships to be elucidated, such as for instance: (i) basal relationships between the NW and OW lineages in the Lupinus phylogeny; (ii) relationships amongst the OW lineages and within the African clade; and (iii) the enigmatic position of some taxa (e.g. Floridian lupines).44,45
Recently, the plastome of L. luteus was published, representing the first chloroplast genome sequenced in Lupinus and in the Genistoid lineage.15 Comparison with other legume plastomes allowed the discovery that the Genistoids share a 36-kb inversion, and the identification of mutational hotspots representing potentially informative regions for evolutionary studies. However, these identified regions, such as the ycf4 gene in the NPAA clade12 or the clpP in the IRLC and Mimosoids,14 may be of interest only in particular clades, due to a specific accelerated evolutionary rate in these lineages. Thus, additional plastomes are needed to specifically understand the plastome evolution of the lupine/Genistoid lineage and to accurately identify their most variable regions that are of phylogenetic significance. In this context, we sequenced four novel lupine plastomes: two Mediterranean smooth-seeded species, L. albus and Lupinus micranthus Guss. and two rough-seeded species, Lupinus atlanticus Glads. and Lupinus princei Harms. Comparative analyses were performed among the five OW lupine plastomes (including the previously published L. luteus15) at the structural (inversions, indels, repeat numbers and distribution), gene and sequence levels, in order to better understand their evolutionary dynamics and to identify novel phylogenetically informative regions. More specifically, sequencing of these four additional plastomes revealed the pseudogenization of the chloroplast rps16 gene in Lupinus species. Analyses of the Ka/Ks ratios of the functional chloroplast and the nuclear rps16 genes (both encoding the same chloroplast RPS16 protein) from some representative Angiosperm species revealed that both copies were under purifying selection. However, since the nuclear rps16 gene also encodes the mitochondrial RPS16 protein and that this gene is lost in the mitochondrial genomes of all plants sequenced to date, the loss of the nuclear rps16 gene would be detrimental for plant survival. This could explain why only the chloroplast rps16 gene has been functionally lost many times during plant evolution, despite being under purifying selection. In addition, investigations on the evolutionary dynamic of the lupine plastomes (mutations, indels and repeated elements) allowed identification of variable characters and regions. The phylogenetic interest of these regions in the genus Lupinus was tested using representative species of the main lupine clades.
2. Material and methods
2.1. Plant material and DNA isolation
Genomic DNA of 30 lupine species was extracted from fresh leaves using the NucleoSpin® Plant II kit (Macherey-Nagel), following the manufacturer’s instructions. The genomic DNA extracts of four Lupinus species (L. albus, L. micranthus, L. atlanticus and L. princei) were subjected to NGS for plastome reconstruction. DNA extracts from the other 26 lupine species were used in different evolutionary tests on genes and regions of interest; including four OW rough-seeded species (L. digitatus, L. cosentinii, L. anatolicus and L. pilosus); three OW smooth-seeded species (L. hispanicus subsp. bicolor, L. angustifolius subsp. angustifolius and L. angustifolius subsp. reticulatus); and fourteen species representing the main known groups in the NW lupines. Among these are (i) five members of the North and South East American clade (L. texensis, L. paraguariensis, L. gibertianus and L. sellowianus); (ii) nine members from various groups mainly occurring in western regions of North, Central and South America (L. affinis, L. hirsitussimus, L. luteolus, L. nanus, L. polyphyllus, L. mutabilis, L. mexicanus, L. elegans, two unidentified samples L. sp. from Equator); (iii) and two Florida endemic species (L. diffusus and L. villosus). Moreover, the DNA of three representatives from the Genista-Cytisus complex, sister group to Lupinus in the Genisteae46 tribe were obtained: Retama sphaerocarpa, Cytisus battandieri and Genista erioclada. More details on geographic origins and reference numbers of these plant materials are presented in Supplementary Table S1.
2.2. High throughput sequencing, plastome assembly and annotation
The genomic DNA of L. albus, L. micranthus, L. atlanticus and L. princei were subjected to high-throughput sequencing using an Illumina HiSeq 2000 platform (BGI, Hong-Kong). One flow cell containing a library of each species was used, yielding ∼11 millions of 100 bp paired-end (PE) reads (insert size = 500 bp) for each library, except L. micranthus, for which ∼5.5 millions of PE reads were obtained.
De novo chloroplast genome (plastomes) assemblies were performed using Paired End Illumina reads and ‘The organelle assembler’ software (http://metabarcoding.org/asm (January 2015, date last accessed))47: its aim is to assemble over represented sequences such as organelle genomes (chloroplast or mitochondrion), or the rDNA cistron. Each draft plastome sequence was then verified and corrected by mapping the Illumina reads against each genome using Bowtie 2 v2.0.48 A few uncertain nucleotides were verified by Sanger sequencing. Plastome annotation was performed using DOGMA (Dual Organellar GenoMe Annotator, http://dogma.ccbb.utexas.edu (January 2015, date last accessed))49 and by aligning each of the four newly constructed plastomes with the published L. luteus plastome (KC69566615). A graphical representation of each plastome was drawn using Circos50 (Supplementary Figs S1–S4).
2.3. Identification of rps16 gene sequences in plant mitochondrion, chloroplast and nuclear genomes
Sequences of the rps16 gene were searched for all non-parasitic plant mitochondrion, chloroplast and nuclear genomes available to date. Organelle and nuclear genomes were downloaded from GenBank (https://ncbi.nlm.nih.gov (November 2016, date last accessed)) and Phytozome v11 (https://phytozome.jgi.doe.gov/pz/portal.html (November 2016, date last accessed)), respectively. For the nuclear rps16 genes, presence in mature proteins of a signal peptide targeting the proteins to the organelles was tested using BaCelLo,51 ProteinProwler,52 TargetP,53 MultiLoc254 and Predotar.55 In addition, as the chloroplast rps16 gene has a subgroup IIB intron, we looked for the presence of the correct splicing of this intron by verifying the presence of the strictly conserved splicing sites (GTGYG and AY at the 5ʹ and 3ʹ splice sites of the intron, respectively)56–59 in all chloroplast rps16 genes with a complete coding sequence (742 species).
2.4. Selective pressure acting on the nuclear and chloroplast rps16 genes
Within a subset of plants representing the main clades of Angiosperms (Arabidopsis lyrata: Brassicales; Citrus sinensis: Sapindales; Cucumis sativus: Cucurbitales; Glycine max: Fabales; Manihot esculentum: Malpighiales; Musa acuminata: Zingiberales; Oryza sativa: Poales; Panicum virgatum: Poales; Prunus persica: Rosales; Solanum lycopersicum: Solanales; Theobroma cacao: Malvales; Vitis vinifera: Vitales), we retrieved the functional nuclear and chloroplast rps16 gene sequences, which both encode the chloroplast RPS16 proteins. The different nuclear or chloroplast copies were aligned using Geneious v6.1.860 and the alignments were adjusted manually. Non-synonymous and synonymous nucleotide substitution rates were evaluated using the yn00 method implemented in PAML61 for the nuclear and chloroplast rps16 sequences. A list of species considered and the accession numbers of nuclear and chloroplast rps16 sequences used are presented in Supplementary Table S2. Ka/Ks analyses of chloroplast rps16 gene were also performed using only the representatives of the following Angiosperm families: Asteraceae, Brassicaceae, Fabaceae, Poaceae and Solanaceae.
2.5. Sequence divergence among lupine plastomes
To identify the most variable regions among lupine chloroplast genomes, the five plastomes were aligned using Geneious v6.1.860 and pairwise comparisons between each of the five plastomes were performed to evaluate the percentage of identity in sliding window frames of 1 kb with a Python custom script. Using this script, insertion-deletions (indels) with a minimum size of 20 bp were identified. These large indels as well as pairwise comparisons results were represented graphically using Circos.50 Additionally, the five aligned plastomes were screened to identify autapomorphous (single) and shared indels of at least 2 bp, and the excluding regions with homopolymers or with ambiguous overlapping indels. Sequence divergence among the five lupine plastomes (including L. luteus) was also evaluated independently for intergenic spacers, introns, exons, rRNAs and tRNAs by calculating pairwise distances between homologous regions. Pairwise distances were determined with the ape R-cran Package62 (available at: http://cran.r-project.org/web/packages/ape/ape.pdf) using the Kimura 2-parameters (K2p) evolution model for introns and intergenic spacers.63 Additionally, sequence divergence of protein encoding sequences was estimated using the synonymous (Ks) and non-synonymous (Ka) nucleotide substitution rates with the yn00 method64 from the PAML package.61
Repeat sequences in each lupine plastome were identified using REPuter65 with similar parameters as previously described for the analysis of Fabaceae plastomes7,15,25,66 and excluding one copy of the IR. Palindrome sequences as well as dispersed direct and IRs of a minimum length of 30 bp and presenting at least 90% sequence identity were identified (Hamming distance of three). Additionally, mono-, di-, tri-, tetra- and penta-nucleotides Short Sequence Repeats (SSRs) with a mininimal size of 12 bp and a minimal repeat number of five were detected using the Phobos software67 implemented in Geneious v6.1.8.60
2.6. PCR amplification of the rps16 gene and the most variable regions in lupines
Several primer pairs were designed using Primer 3.068 to examine the variation of the rps16 gene and two fast evolving regions (psaA-ycf4 and ycf1-rps15) in Lupinus. For rps16, the following primer pairs were used: F-CCGTCCCAGAGCATATTCAG, R-GCAACGATTCGATAAATGGC and F-CCCATTCATATCGAAGGAAAACT, R-CCATCATGTACTATTTACATCATCAATC and R-CTATATACAAGTCATCCACACCCTC. Within the fast evolving regions, primer pairs were designed to amplify four sub-regions (accD and ycf1 genes, ycf1-rps15 and trnFGAA-trnLUAA 5ʹ-3ʹ intergenic spacers): accD with F-GTCTATAAATACATTACCCCCG, R-TGTCTTCATCCATAGGATTCC; ycf1-rps15 with F-GATTTATGTTGCACAAACCG, R-CATTGATGGGTGGTGAGG; trnFGAA-trnLUAA with F-TTGAACTGGTGACACGAGG, R-TGGCGAAATTGGTAGACG. Because of the large size of the ycf1, two primer pairs were designed: ycf1 part1 with F-AATCAAGCAGAAAGTTATGGG, R-CTTACATCTTTTGAGCTTTCACTC; ycf1 part2 with F-GGAATGGAAGTAGAATTGCC, R-TTTTGTTTACGCGTCTTGT.
PCR amplifications of these regions were carried out for 32 taxa (including three Genisteae outgroups, Supplementary Table S1) in a total volume of 50 μl, containing 5× Green GoTaq flexi Reaction Buffer (Promega), 0.2 mM of dNTP, 0.2 μM of each primer, 1.25 Unit of G2 flexi DNA polymerase (Promega), mqH2O and 20 ng of template DNA. Cycling conditions were 94 °C for 2 min followed by 35 cycles at 94 °C for 30 s, 48–52 °C (adapted according to the primer pairs used) for 30 s, 72 °C for 90 s and a final extension at 72 °C for 7 min. PCR products were purified using the NucleoSpin gel and PCR clean up kit (Macherey Nagel), following the manufacturer’s instructions. Purified PCR products were sequenced directly (in both directions) by Sanger at Macrogen Europe (Amsterdam, The Netherlands). All sequences were deposited in Genbank under the accession numbers, KX147685 to KX147753 and KX787895 to KX787910.
2.7. Phylogenetic analyses
For each of the four chloroplast regions investigated, all lupine sequences were aligned using MAFFT implemented in Geneious v6.1.8.60,69 The resulting alignments were adjusted manually. In addition, a concatenated data matrix was constructed using the sequences obtained from the four regions (accD and ycf1 genes, ycf1-rps15 and trnFGAA-trnLUAA 5ʹ-3ʹ intergenic spacers). These matrices were first subjected to phylogenetic analyses using Maximum Parsimony (MP). Bootstrap analyses were performed with 1,000 replicates.70 These data matrices were also subjected to Maximum Likelihood (ML) phylogenetic analyses. The best-fitted model of sequence evolution for each region (individual or concatenated) was determined using JModeltest71 and ML analyses were then performed for each matrix with 1,000 bootstrap replicates using MEGA 6.0.72
3. Results and discussion
3.1. Structure, organization and gene content of lupine plastomes
The Illumina PE reads obtained for L. albus, L. atlanticus, L. micranthus and L. princei, were used to assemble their chloroplast genome sequences (deposited in GenBank under accession numbers KU726826; KU726827; KU726828; KU726829, respectively). The four plastomes harbor a quadripartite structure (a Large Single Copy and a Small Single Copy separated by two IRs) with a total length ranging from 151,808 bp to 152,272 bp. As expected from previous PCR-based evidence, they all have the 36-kb inversion that occurred at the base of the Genistoid emergence or soon after.15 The different Lupinus plastomes have similar gene, intron and GC content (Table 1) as do most photosynthetic and non-parasitic angiosperm plastomes.11 The genes are distributed into three main categories: self-replication (58 genes), photosynthesis (47 genes) and other functions (six genes) (Supplementary Table S3). Among these genes, 76 are protein-encoding genes, 30 encode tRNAs and four encode rRNAs. None of the genes known to be lost or pseudogenized in other legume lineages, such as accD,7psaI, ycf4, rpl23 or rpl3312 are missing in the lupine plastomes. Interestingly, comparative analyses of the lupine plastomes (including L. luteus) revealed a likely loss of functionality of the rps16 gene in L. albus and L. micranthus but not in the other species. Both pseudogenes showed a deletion (verified by Sanger sequencing), which lead to a pre-mature stop codon (19 and 20 amino acids earlier in L. albus and L. micranthus, respectively) within the functional domain of the RPS16 protein (Fig. 1). To determine if these truncated rps16 genes in L. albus and L. micranthus are still functional, we used pfam (pfam-A, default parameters)73 to search for the presence of a functional domain in the five lupine species investigated in this study. No RPS16 functional domain could be identified in L. albus and L. micranthus only, clearly suggesting that rps16 is a pseudogene in these two lupine species. Recently, an additional way at the origin of the loss of functionality of the chloroplast rps16 gene was identified and corresponds to the loss of its splicing capacity.59 In lupines, we observed that the rps16 intron is not correctly spliced. This suggests that the rps16 is not functional in the five chloroplast genomes (all five lupine plastomes must therefore have 76 functional protein-coding genes), and that the loss of functionality most likely occurred first via the loss of the ability to splice the intron. Thereafter, additional mutations in L. albus and L. micranthus led to pre-mature stop codons. Whether this shared pre-mature stop codon results from a common ancestor or from independent mutational events needs to more accurately resolve phylogenetic relationships of these two species among the OW lupines (see later in the phylogenetic section). Sequencing of the rps16 gene in other lupines and closely related species revealed that another population of L. micranthus has a pseudo rps16, and that it is also defunct in L. angustifolius, Lupinus mariae-josephae, L. villosus and in a member of the Lupinus sister group, G. erioclada (data not shown).
Table 1.
Characteristics of Lupinus plastomes
| Plastome characteristics | L. luteus | L. albus | L. atlanticus | L. princei | L. micranthus |
|---|---|---|---|---|---|
| Overall size in bp | 151,894 | 151,921 | 152,272 | 152,243 | 151,808 |
| LSC size in bp (%) | 82,327 (54.2) | 82,280 (54.2) | 82,674 (54.3) | 82,663 (54.3) | 82,145 (54.1) |
| SSC size in bp (%) | 17,847 (11.7) | 17,841 (11.7) | 17,894 (11.8) | 17,876 (11.7) | 17,857 (11.8) |
| IR size in bp (%) | 25,860 (34.1) | 25,900 (34.1) | 25,852 (34) | 25,852 (34) | 25,903 (34.1) |
| Coding regions size in bp (%) | 90,217 (59.4) | 90,002 (59.2) | 90,125 (59.2) | 90,104 (59.2) | 90,083 (59.3) |
| Protein-coding region in bp (%) | 78,363 (51.6) | 78,148 (51.4) | 78,271 (51.4) | 78,250 (51.4) | 78,229 (51.5) |
| Introns size in bp (%) | 19,136 (12.6) | 19,115 (12.6) | 19,111 (12.6) | 19,121 (12.6) | 18,754 (12.4) |
| rRNA size in bp (%) | 9,056 (6) | 9,056 (6) | 9,056 (5.9) | 9,056 (5.9) | 9,056 (6) |
| tRNA size in bp (%) | 2,798 (1.8) | 2,798 (1.8) | 2,798 (1.8) | 2,798 (1.8) | 2,798 (1.8) |
| IGSs size in bp (%) | 42,541 (28) | 42,804 (28.2) | 43,036 (28.3) | 43,018 (28.3) | 42,971 (28.3) |
| No. of different genes | 110 | 110 | 110 | 110 | 110 |
| No. of different protein-coding genes | 76 | 76 | 76 | 76 | 76 |
| No. of different rRNA genes | 4 | 4 | 4 | 4 | 4 |
| tRNA genes | 30 | 30 | 30 | 30 | 30 |
| No. of different duplicated genes by IR | 17 | 17 | 17 | 17 | 17 |
| No. of different genes with introns | 18 | 18 | 18 | 18 | 18 |
| Overall % of GC content | 36.6 | 36.7 | 36.6 | 36.7 | 36.6 |
| % of GC content in protein-coding regions | 37.3 | 37.3 | 37.3 | 37.3 | 37.3 |
| % of GC content in introns | 36.3 | 36.9 | 36.8 | 36.8 | 36.8 |
| % of GC content in IGSs | 30.3 | 30.4 | 30.3 | 30.4 | 30.3 |
| % of GC content in rRNA | 55.3 | 55.3 | 55.3 | 55.3 | 55.3 |
| % of GC content in tRNA | 53.3 | 53.2 | 53.3 | 53.3 | 53.3 |
Figure 1.
Comparison of lupine chloroplast rps16 coding-sequences with legume rps16 sequences (Glycine max and Lotus japonicus) and Cucumis sativus rps16 sequence (outgroup). The ribosomal protein S16 domain is indicated between brackets. The presence of a pre-mature stop codon within the rps16 functional domain of L. albus and L. micranthus is represented by a black asterisk. The black triangle denotes the position of rps16 intron. It is important to note that the five lupine species present incorrect splicing sites according to.59
3.2. Evolutionary dynamic and fate of the rps16 gene in plant mitochondrial, chloroplast and nuclear genomes
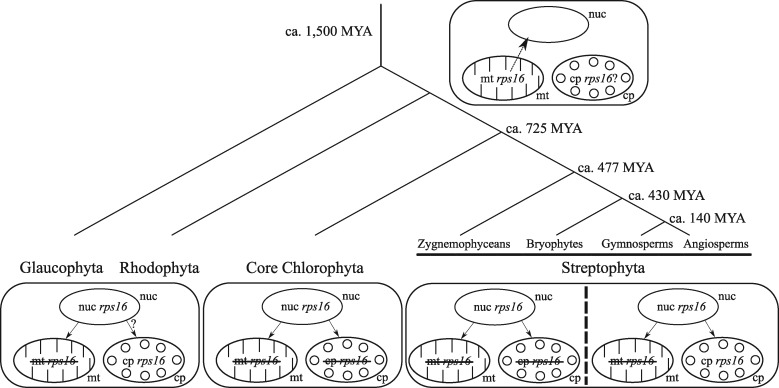
As in some Lupinus species, the chloroplast rps16 gene was missing in many other Fabaceae, including P. vulgaris and the IRLC.12 In this family, the chloroplast rps16 gene, which is essential for plant survival, has been functionally replaced by a nuclear gene that can encode both mitochondrial and chloroplast RPS16 proteins.28 To better understand the origin and evolutionary fate of rps16 genes residing in different genome compartments but with similar functions, we searched for plant chloroplast, mitochondrial and nuclear rps16 genes in the currently available nuclear and organelle genomes. In total, we investigated 52 nuclear, 289 mitochondrion and 1,166 chloroplast genomes from the non-parasitic brown and green plant lineages (Haptophytes, Stramenopiles, Glaucophytes, Rhodophytes, Chlorophytes and Streptophytes). Within all the sequenced mitochondrial genomes, no rps16 gene was found, whereas a functional (no pre-mature stop codon) nuclear rps16 gene copy was observed in all species investigated. The loss of the mitochondrial rps16 gene before the divergence of the Glaucophyta from Rhodophyta, Chlorophyta and Streptophyta lineages suggests that the transfer of rps16 from the mitochondrion to the nucleus occurred ∼1,500 million years ago74 (Fig. 2), which is much earlier than previously determined (i.e. before the emergence of angiosperms).28 As the rps16 gene was lost from the mitochondrion before divergence of the green lineage and as the nuclear rps16 gene encodes mitochondrion RPS16 proteins, its functional loss from the nuclear genome would be detrimental. As expected, a peptide signal targeting the nuclear encoded RPS16 proteins to the mitochondrion was identified in all species investigated, while the presence of a chloroplast target peptide was predicted in only a few species (Supplementary Table S4). However, it is likely that all species present a nuclear rps16 gene that can target the protein to both the mitochondria and plastids, as previously demonstrated by.28 Indeed, these authors showed experimentally that in two species that have lost the rps16 gene from their chloroplast (Medicago truncatula and Populus alba), and for which the nuclear encoded RPS16 proteins were only predicted to be localized in the mitochondrion (Supplementary Table S4), the RPS16 proteins were targeted to both organelles. It is thus very likely that despite the absence of a predicted chloroplast target peptide, nuclear-encoded RPS16 proteins are targeted to both mitochondria and chloroplasts.
Figure 2.
Genome localization of the rps16 gene(s) encoding the mitochondrial and chloroplast RPS16 proteins in Archeaplastida. Early in Archeaplastida, the mitochondrial rps16 gene was transferred to the nucleus (nuc) and acquired a signal peptide targeting both mitochondrion (mt) and chloroplast (cp). In Glaucophytes and Red Algae, the mitochondrial rps16 gene is always absent whereas it is present in 13 Rhodophyta chloroplast genomes (no plastome sequence available from Glaucophytes). In the core Chlorophyta lineage, none of the 76 species having a fully sequenced chloroplast and mitochondrial genomes have a rps16 gene. In the Streptophyta lineage, no rps16 gene was found in the mitochondrial genomes, whereas the chloroplast rps16 gene may either be functional or loss its functionality (complete gene loss, presence of a pre-mature stop codon or loss of the splicing capacity). Tree was redrawn according to Ref.97,98
Within the chloroplast genomes investigated (1,166), the rps16 gene was found to be missing (total absence of the gene or truncated proteins due to pre-mature stop codon) in 312 genomes. We looked for the presence of correct splicing sites in chloroplast rps16 genes with a complete coding sequence and an intron. We found that 434 and 306 had or did not have the splicing capacity, respectively. Among the latter, 197 exhibited mutations in both 5ʹ and 3ʹ splice sites, whereas 22 and 87 had mutations only at the 5ʹ or the 3ʹ splice sites, respectively (Supplementary Table S5). As previously observed, this gene has lost its functionality many times during flowering plant evolution,12 by the loss of either all or part of the coding sequence or of the splicing sites. Dating back to 1,500 MYA, chloroplast RPS16 proteins can be produced by either nuclear or chloroplast rps16 genes. Our results highlight the fact that even though the chloroplast rps16 gene could have been non-functional in all plant genomes since then, it is still present and functional in most plants.
To better understand the evolutionary dynamics of the rps16 gene, we analysed the selective pressure acting on functional chloroplast and nuclear rps16 genes among 12 species representing the main angiosperms clades. As the rps16 gene was functional in both chloroplast and nuclear genomes and as only the chloroplast copy is likely to be lost, the selective constraints acting on the chloroplast gene could be relaxed. Results of Ka/Ks ratios revealed a strong purifying selection for both chloroplast and nuclear rps16 (average Ka/Ks ratio: 0.045 ± se 0.010 and 0.1707 ± se 0.003 for nuclear and chloroplast copies, respectively; Supplementary Tables S6 and S7). Ka/Ks ratios of cp-rps16 for each of the main Angiosperm families (Asteraceae, Brassicaceae, Fabaceae, Poaceae and Solanaceae) were also calculated for all cp-rps16 found with a complete coding sequence and a correctly spliced intron. Results were similar for each of the five families investigated and revealed a strong purifying selection acting on all the tested datasets (average of Ka/Ks ratios always lower than 0.23; Supplementary Table S8). A possible explanation of this strong negative selection pressure still acting on the chloroplast rps16 gene and the presence of a functional chloroplast rps16 gene in many plant genomes (in contrast with plant mitochondrial genomes) is that the chloroplast rps16 gene may function or be regulated slightly better than the nuclear gene under certain conditions.
Although these results revealed the multiple status (absent, truncated, incorrectly spliced, functional) of the chloroplast rps16 gene among the plant kingdom and confirmed the hypothesis of59 that the loss of splicing capacity is widely spread through plant species, mechanisms beyond the conservation of the chloroplast copy in most species remain unknown. Indeed, the chloroplast rps16 gene is still essential in certain plant species as revealed by knock-down studies in tobacco.29 Different hypotheses have been proposed to explain the retention of some genes within the organelle genomes. The current most widely accepted hypothesis corresponds to the Colocation of gene and gene product for redox regulation of gene expression (or CoRR).75–77 This hypothesis concerns genes that are redox-dependant (such as rbcL; rps2,3,4,7,8,11,12,14,19; rpl2,14,16,20,36).77 However, the chloroplast rps16 gene has been found to be redox independent.78 An alternative hypothesis that was considered concerned the retention of the ribosomal assembly genes in the organelle.79 A ‘core set’ of ribosomal genes were identified in all plants investigated,79 however, rps16 was not included. Another possible explanation of the retention of a functional chloroplast rps16 gene in many species may be due to the loss of the chloroplast target peptide of the nuclear-encoded RPS16 protein (despite the fact that the mitochondrion rps16 target signal remain retained).
3.3. Lupine plastome variability
3.3.1. Identification of single nucleotide polymorphims and indels in lupine plastomes
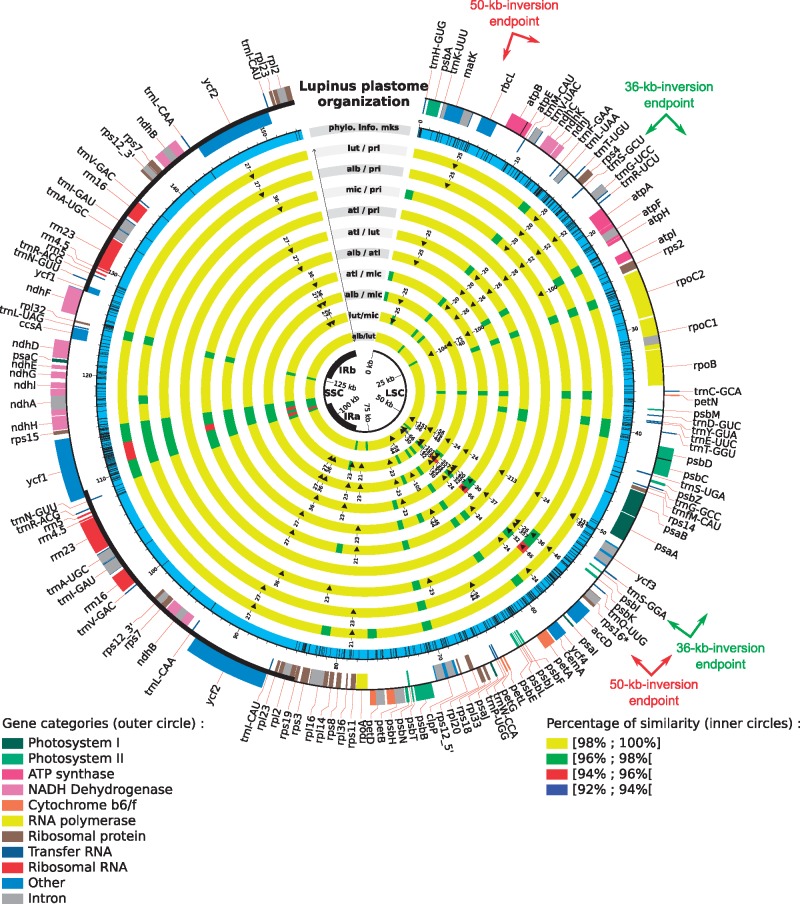
To identify putative mutation hotspots, pairwise comparisons of the five lupine plastomes were performed and showed that they have a very high level of sequence identity (98% on average). The two African species (L. atlanticus and L. princei), which are the most closely related lupine species investigated in this study, exhibit the highest identity (99.7%); whereas the species with the lowest sequence identity are L. luteus and L. micranthus (97.9%). These comparisons also enabled identification of 164 non-ambiguous indels along the chloroplast genomes, including 14 with a size ranging from 20 to 357 bp. Of the 164 indels, 50% are 5–6 bp long. These analyses revealed two highly variable regions (Fig. 3). The first region spans from psaA to ycf4 (∼11.5 kb) and was already identified as a hypermutable region.12 This region contains 11 genes: psaA, ycf4, ycf3, trnS-GGA, psbI, psbK, trnQ-UUG, rps16, accD and psaI genes, for which four genes (accD, rps16, ycf4, psaI) were shown to be functionally lost in at least one legume species. The second most variable region includes the ycf1-rps15 genes (∼6.5 kb). The ycf1 gene, which encodes a translocon protein of the inner chloroplast membrane,80 is larger than 5 kb in lupines and is highly variable with the exception of a 5ʹ fragment duplicated in the IR (519 bp in lupines). This gene was recently identified as one of the most variable chloroplast genes in Angiosperms and is considered as a powerful tool for DNA barcoding.81,82 The longest hypervariable region (psaA-ycf4) contains the highest number of indels (25), with 11 (among the 14 present in the genome) between 20 and 357 bp. Some of these large indels will most likely be useful to discriminate lupine species and/or groups of species from other Lupinus lineages or from other closely related genera.
Figure 3.
Pairwise comparison of lupine plastomes to identify single nucleotide polymorphisms and indels. The outer circle represents the gene map of lupine plastomes; the boxes outside this first circle indicate a counterclockwise of transcription direction whereas inside boxes indicate a clockwise transcript direction. In the second circle, potentially informative sites are indicated by black bars. The following ten inner circles represent pairwise comparisons between the five available lupine plastomes; pairwise identity level is indicated and indels >20 bp are represented by black triangle. The central black circle represents the different parts of the chloroplast genome (LSC, SSC and IRs). The endpoints of the 50-kb inversion, specific to the Papilionoid legumes20–22 and of the 36-kb inversion, specific to the Genistoid clade,15 are represented by arrows.
3.3.2. Sequence divergence between lupine plastomes
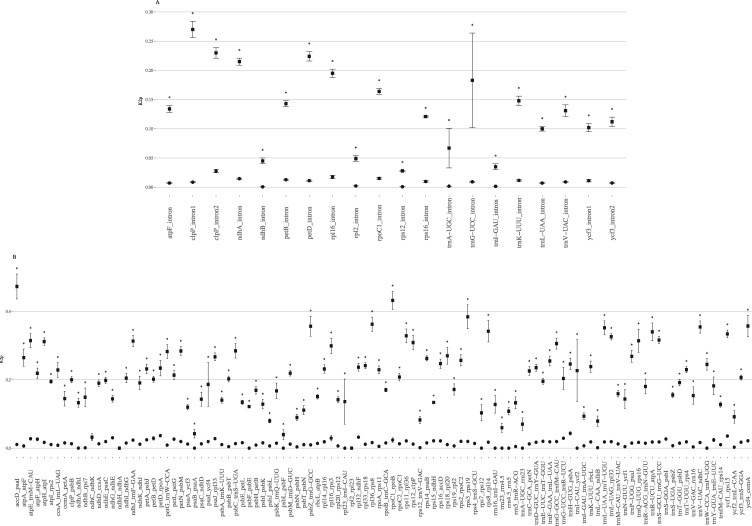
Pairwise distance (K2p) comparisons among the five lupine plastomes were calculated for non-coding sequences. As expected, the lowest rates of variation were observed for tRNA and rRNA (maximum K2p value: 0.0141, Supplementary Table S9). For introns (Supplementary Table S9, Supplementary Fig. S6A), average of K2p rates ranged from 0.0006 (ndhB intron) to 0.0263 (clpP intron 1). Compared with the K2p analyses performed by,15 who estimated sequence divergence between L. luteus and other legume species, our overall K2p values obtained by comparing only lupine species are, as expected, significantly lower (Wilcoxon test, P-value = 0.05; see Fig. 4A). Among the five lupine species considered, the clpP intron 1, rpl16, rpoC1 and ndhA introns exhibit the higher K2p values. The most variable intron in lupines corresponds to the first intron of clpP, which also showed accelerated mutation rate in Mimosoideae.14,18 The trnK and trnL introns previously used for legumes and lupines phylogenies were found to only exhibit high variation when comparing L. luteus to other Fabaceae.
Figure 4.
K2p mean values ± standard error for (A) introns and (B) intergenic spacers between (i) L. albus, L. atlanticus, L. luteus, L. micranthus and L. princei (black cicles) and (ii) L. luteus, Phaseolus vulgaris, Pisum sativum, Vigna radiata, Glycine max, Lathyrus sativus, Cicer arietinum, Trifolium subterraneum, Medicago truncatula, Lotus japonicus and Millettia pinnata (black squares).15 The x-axis corresponds to intron and intergenic regions. Asterisks represent a statistically significant difference (P.value: 0.05).
K2p values for IGSs ranged from 0 to 0.0434 (Supplementary Table S9, Supplementary Fig. S6B). In comparison to the commonly-used IGSs in legume phylogenetic studies (trnF_trnL, mean K2p = 0.0185, 428 bp; trnL_trnT, 0.0182, 633 bp; trnS_trnG, 0.0181, 799 bp), 36 IGS regions present a higher K2p values, and 15 of them are larger than 300 bp, and thus may be useful for phylogenetic studies. This analysis allowed detection of two relatively variable IGS sequences, corresponding to ycf1_rps15 (mean K2p = 0.0355, aligned length = 470 bp) and rpl32_ndhF (0.0322, 486 bp) that were not detected in previous analysis (Fig. 4B).
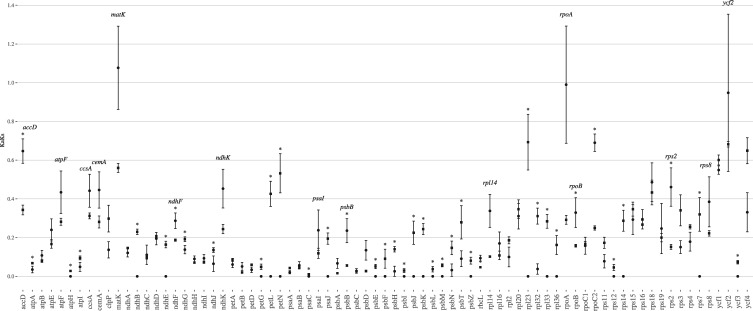
Non-synonymous (Ka) and synonymous (Ks) nucleotide substitution rates were calculated for protein-coding sequences, as well as the Ka/Ks ratio (Supplementary Tables S10–S12). The mean Ks among the five lupines studied ranged from 0 (petG, petL, petN, psaJ, psbF, psbI, psbM, rpl23, rpl33, rps7) to 0.05049 (psbT). All protein-encoding genes have a Ks value lower than 0.1. Similarly, the non-synonymous substitution rate (Ka) was lower than 0.025 for all genes. Finally, Ka/Ks ratios were calculated for each protein-coding region in order to determine the selective constraint acting on each gene. Almost all genes evolved under high purifying selective constraint (53 of the 77 genes have a Ka/Ks ratio lower than 0.2), except for six genes (matK, rpoA, ycf2, rpoC2, accD and ycf1) that show a ratio > 0.5 (including three genes evolving almost neutrally (matK, rpoA, ycf2; Supplementary Fig. S7). Except for ycf1 and ycf2, the other genes were not identified as neutrally evolving between legumes and L. luteus.15 Comparison of Ka/Ks ratios obtained when considering only lupine species to the Ka/Ks ratios obtained when comparing L. luteus15 to other legumes, revealed 14 genes that exhibit higher Ka/Ks ratios between lupines than between L. luteus and legumes. Among these genes, only six are significantly higher (accD, ndhF, psbB, rbcL, rpoB and rsp2; Fig. 5). However, detailed analysis of these genes (synonymous and non-synonymous substitution comparisons; and codon-based ML phylogenetic analyses; results not shown) did not reveal significant accelerated mutation rates at either synonymous or non-synonymous sites. Conversely, the ycf4 gene and the flanking cemA and accD genes, which were found to be highly variable in the Lathyrus and Desmodium clades,12 were more stable, lacking major rearrangements in Lupinus. These results highlight that fast-evolving regions may strongly differ among clades within a family.
Figure 5.
Mean Ka/Ks ratio values ± standard error between homologous regions of (i) the five Lupines L. albus, L. atlanticus, L. luteus, L. micranthus and L. princei (black circles) and (ii) L. luteus, Phaseolus vulgaris, Pisum sativum, Vigna radiata, Glycine max, Lathyrus sativus, Cicer arietinum, Trifolium subterraneum, Medicago truncatula, Lotus japonicus and Millettia pinnata (black squares15). The x-axis corresponds to the CDS regions. Asterisks represent a statistically significant difference (P.value: 0.05).
3.3.3. Lupinus plastid sequences of phylogenetic utility
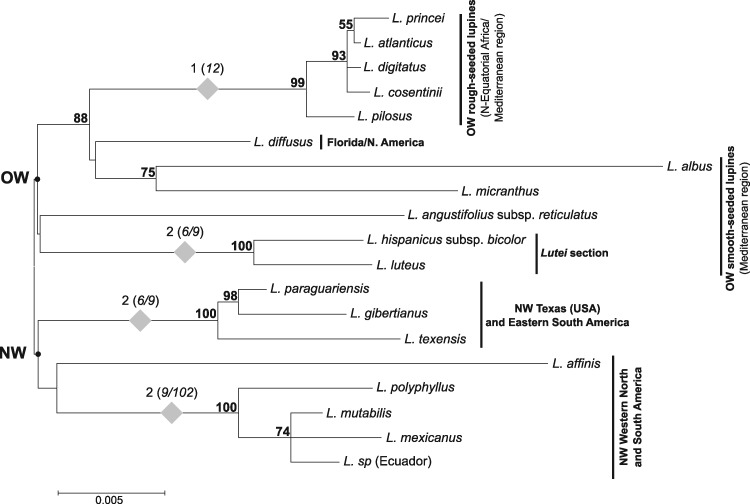
To explore the putative phylogenetic utility of different chloroplast regions, potentially informative sites (Pi) were evaluated among lupines in: (i) complete chloroplast genomes, (ii) protein-coding sequences, (iii) intergenic spacers, (iv) introns and (v) in the two hypervariable regions (psaA-ycf4 and ycf1-rps15) (Table 2). Results revealed 666 Pi (among 2,874 variables sites) in the five aligned plastomes, which are distributed as follow: 45.3% of the Pi in IGS, 38.3% in CDS and only 11.4% in introns. The remaining five percent are located in tRNA and rRNA genes. The two hypervariable regions containing psaA-ycf4 and ycf1-rps15, account for 8.7 and 14.3% of the total number of Pi, respectively. Molecular phylogenies were performed using either complete plastomes, or introns, or IGS or CDS, and revealed a similar topology (Supplementary Fig. S5), with the rough-seeded species (L. atlanticus and L. princei) in a well-supported clade (always with 100% of bootstrap support) clearly distinct from the smooth-seeded species L. albus, L. luteus and L. micranthus. Among the latter, L. albus was always the closest Mediterranean lupine to the rough-seeded group (86–87% of bootstrap value based on either IGS or complete plastome data). These results differ from previous phylogenies40,41,44,45 based on single or few genes (chloroplast and nuclear genes), which found L. micranthus to be the closest Mediterranean lupine to the rough-seeded species. In addition, the whole plastome phylogenies provide, for the first time, strong evidence (97–99% bootstrap support) of a common ancestor for L. micranthus, L. albus and the rough-seeded lupines, which are positioned as sister group to L. luteus. MP analyses (using PAUP483) of the five aligned lupine plastomes, with or without the 164 non-ambiguous indels (coded as 0 or 1 for the presence of a deletion or an insertion, respectively), led to the same results (not shown). To further investigate the phylogenetic utility of the most variable lupine regions identified, we amplified and sequenced five chloroplast regions (accD, two parts of the ycf1 gene and the ycf1-rps15 and trnFGAA-trnLUAA 5ʹ-3ʹ intergenic spacers, length ranging from 800 to 2,000 bp) from 16 lupine species. Each matrix was subjected to ML analysis. After verifying the absence of incongruence between the trees obtained for the five different regions, a concatenated matrix of all regions was analysed, following the conditional-combination approach.84L. villosus and L. anatolicus were not considered in this analysis, as not all regions were amplified in these two species. The ML tree obtained from this latter matrix is presented in Fig. 6. Despite low resolution of the basal nodes, the topology is consistent with an early divergence of the lupines into two main lineages: the OW lineage comprising all the smooth- and rough-seeded Mediterranean and African taxa, which includes the representative of the Floridian species (L. diffusus) and the NW lineage composed of all American taxa from diverse origins (except L. diffusus). Within these two main lineages, most clades are consistent with previous phylogenies40,41,44,45 and some of them present very high support using these cpDNA data, such as: (i) the OW rough-seeded species (L. atlanticus, L. cosentinii, L. digitatus, L. pilosus and L. princei) with a bootstrap value of 99%; (ii) the Mediterranean smooth-seeded lupines L. luteus and L. hispanicus subsp. bicolor (which together form the lutei section) with 100% bootstrap support, linked to L. angustifolius as sister group; (iii) the clade including the Texan lupines and the eastern South American species (L. texensis, L. paraguariensis and L. gibertianus) with 100% bootstrap support, (iv) and a clade (100% bootstrap support) corresponding to the Western American and Mexican species (L. polyphyllus, L. mutabilis, L. mexicanus and the undetermined lupine from Ecuador). Support for these clades is reinforced by synapomorphic indels (Fig. 6). Within the OW lineage, the Mediterranean smooth-seeded species do not form a distinct clade and appear as paraphyletic to the rough-seeded group and the Floridian L. diffusus, L. albus and L. micranthus are placed (with 88% bootstrap support) as the closest Mediterranean smooth-seeded lupines to the rough-seeded species. In this phylogeny, L. albus is sister to L. micranthus, with moderate bootstrap support (75%) rather than to the rough-seeded lupins (with a bootstrap support of 86–87%), as observed in the whole plastome based phylogenies (see above and Supplementary Fig. S5). This incongruence might be explained by the low number of taxa or to the different sequence datasets analysed in these phylogenies (Fig. 6, Supplementary Fig. S5). Further investigation to resolve such phylogenetic uncertainty is needed. Compared with previous Lupinus phylogenies, based on chloroplast (matK, rbcL, trnL intron, trnL-trnF, trnS-trnG, trnT-trnL) or nuclear sequences (LEGCYC1A, LEGCYC1B, ITS1 + 2, GPAT1, GPAT2, SymRK, ETS),39,40,42,44,85 our five hypervariable chloroplast regions may not have revealed novel relationships but strongly reinforced support for some known clades (such as the West and the East American groups, or the OW rough-seeded section). Moreover, they provided additional and significant data supporting the singular Floridian unifoliolate lupines (represented here by L. diffusus), for which phylogenetic placement has always been questionable, as close relative to the OW lupines rather than to the NW ones (at least from the maternally inherited plastome). Finally, we showed the phylogenetic utility of these two identified regions but the consideration of a higher number of lupines and related species will allow for optimal exploitation of their potential to inform these phylogenies, and improve our knowledge of the evolutionary history of lupine and closely related Genistoid clades that are poorly investigated.
Table 2.
Number of potentially informative sites in complete plastomes (cp), protein-coding sequences (CDS), intergenic spacers (IGS), introns as well as in the two hypervariable regions
| Regions | Complete cp | CDS | IGS | Introns | tRNA-rRNA | psaA-ycf4 | ycf1-rps15 |
|---|---|---|---|---|---|---|---|
| Number of Pi | 666 | 255 | 302 | 76 | 33 | 58 | 95 |
| % of total Pi | 100 | 38.3 | 45.3 | 11.4 | 5 | 8.7 | 14.3 |
| Aligned length | 153,462 | 76,518 | 37,948 | 16,093 | 22,903 | 11,534 | 6,092 |
| % of Pi by region | 0.4 | 0.3 | 0.7 | 0.5 | 0.1 | 0.5 | 1.6 |
Figure 6.
Maximum likelihood unrooted tree (General Time Reversible model, rates Gamma distributed with Invariant Sites, 1,000 bootstraps) of concatenated regions (part of accD and ycf1 genes, ycf1-rps15 IGS and trnF-trnL regions). Bootstrap support values are indicated above branches. Grey diamonds represent indels specific to a node. The numbers above the diamonds indicate the number of additional indels supporting the node, with corresponding indel sizes (in bp) between brackets. The Old Wold (OW) and New World (NW) ancestral nodes are indicated on the tree by solid black points.
3.3.4. Repeated sequences
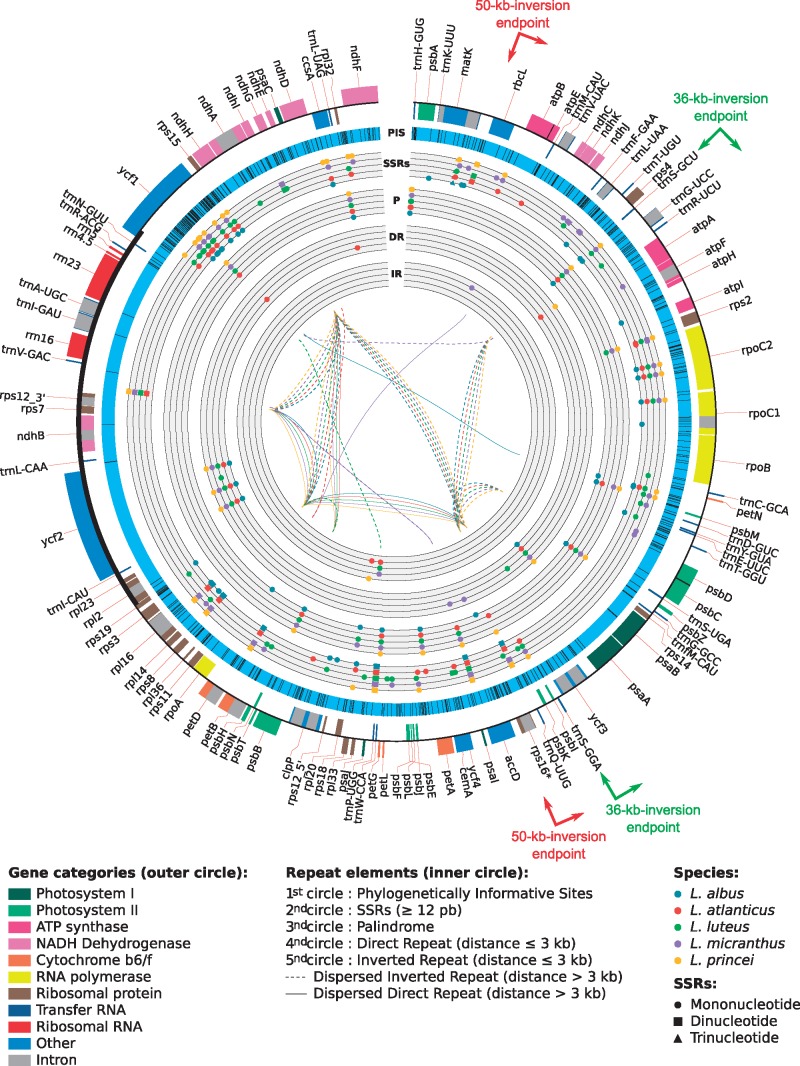
Repeated sequences are known to play a major role in genome evolution. In chloroplast genomes repeats are involved in various structural rearrangements, such as inversions, insertions or deletions. These structural modifications sometimes lead to pseudogenization or duplication as well as to plastome expansion or contraction.10,14,19,86–88 The most striking example of the involvement of repeat sequences in genome size change was observed in Geraniaceae, where the plastome size varies from 128,787 bp to 217,942 bp in Monsonia speciosa and Pelargonium hortorum species, respectively.9 In Fabaceae, repeat sequences were also shown to be involved in LSC extension in Mimosoids14 or related to structural rearrangements, as in Trifolium subterraneum that presents numerous reorganization events and a very high percentage of repeated elements (20% of its genome). Recently, a 29 bp IR in the trnSGGA and trnSGCU was found to be at the origin of a large 36 kb inversion discovered in L. luteus (and Genisteae), through a flip-flop recombination event.15 This inversion was regarded as a new powerful clade marker for most Genistoids in legumes and our study confirmed the presence of this inversion in the four additional Lupinus plastomes investigated here. Since these short inverted repeats (separated by at least 30 kb) are present in almost all known Fabaceae plastomes,15 it has been underlined that such inversion events could have occurred and could occur again elsewhere via the same mechanism.15 Interestingly,16 recently discovered an independent 39 kb inversion at exactly the same location in Robinia pseudoacacia among 13 taxa investigated. This result confirms the potential of such repeats in plastome dynamics, and demonstrates that even rarely occurring, large inversions might result from independent events in distantly related taxa, such as here in Robinioids and Genistoids, biasing their phylogenetic utility. Despite the homoplasious nature of these inversions, such remarkable parallel inversions could be cautiously used as clade evolutionary markers in each of the affected lineages. Because of the importance of repeated elements in plastome evolution (particularly in Fabaceae), we investigated the type and number of repeats present in each Lupinus plastome using REPuter.65 A total of 142 repeats were identified across the five Lupinus species. These repeats, which are relatively well distributed along the plastome sequences (Fig. 7), were divided into three categories, (i) palindromes (60 repeats), (ii) forward repeats (45 repeats) and (iii) reverse repeats (37 repeats). Although all five chloroplast genomes show a relatively similar number of repeats (24 in L. albus to 33 in L. princei) and confirm previous results obtained by15, we identified three, four, nine and six repeats specific to L. atlanticus, L. albus, L. micranthus and L. luteus, respectively.
Figure 7.
Distribution of repeated sequences and potentially informative SNPs in lupine plastomes. From the outer to the most inner circles. First circle: representation of genes content; second circle: potentially informative sites; third circle: SSRs (circles correspond to mononucleotides, squares stand for dinucleotides and triangles represent trinucleotides); fourth circle: direct repeat interspaced by < 3 kb; fifth circle: inverted repeat interspaced by < 3 kb; sixth circle: palindromic repeats. In the middle, full and dotted lines represent direct and inverted dispersed repeats (separated by > 3 kb), respectively. The endpoints of the 50-kb inversion, specific to the Papilionoid legumes20–22 and of the 36-kb inversion, specific to the Genistoid clade,15 are represented arrows.
The number of repeats found across lupine plastomes is much lower than the number observed in some other legumes, such as in T. subterraneum, M. truncatula, G. max, Pisum sativum, or Lathyrus sativus and Cicer arietinum which contain ∼500, 190, 100, 74, 78 and 75 repeats of a similar size, respectively.17,18,66 It is thus not surprising to observe a more conserved genome size, gene content and less structural rearrangement in the Lupinus genus. Distribution of these repeats (mononucleotides, dispersed and palindromic) along plastomes was characterized, revealing that around 70% of the repeats are in the LSC, whereas 20% and 10% are localized in the SSC and IR, respectively. Within the plastome, most of the repeats are situated in the highly variable regions of the LSC, in the rps12-trnVGAC intergenic spacer of the IR and in the intron of ndhA in the SSC. The five lupine plastomes exhibit a similar pattern of repeat distribution, with more than half of the repeats localized in the non-coding regions, around 30% are in protein-coding sequences and around 15% are in introns. Only two dispersed repeats (shared by all lupines) were found in tRNA genes, including the inverted repeat found to be involved in the 36-kb inversion.15 While performing these analyses, we also paid particular attention to Simple Sequence Repeats (SSRs or microsatellites) that are particularly interesting in a wide range of genetic studies in population genetics, plant evolution and domestication, or for the estimation of gene and pollen flow.89–96 We found between 37 (L. princei) and 51 (L. luteus) microsatellites longer than 12 bp in lupine plastomes, with mononucleotide repeats representing between 70 and 80% of these microsatellites, compared with only 16–30% and 0–5%, of di- and tri-nucleotide SSRs, respectively. The ycf1 gene, which corresponds to the most variable lupine plastome regions, is the richest region in SSRs, with ∼20% of the total microsatellites. In comparison, the second hypervariable region (psaA-ycf4) presents only zero to five percent of SSRs (Supplementary Table S13). Among the 213 SSRs identified within the five plastomes, nine are perfectly shared by all species, two additional shared SSRs vary in size, whereas the others are species or group-specific and thus represent potentially useful markers. Taken together, the various kinds of markers revealed from this study (Single Nucleotide Polymorphisms or SNPs, indels, repeats and inversions) represent important resources of genetic/genomic markers with which to deepen our investigations of Lupinus and its Genistoid allies, and for comparative analyses in legumes.
4. Conclusion and further perspectives
In this work, four additional lupine chloroplast genomes were sequenced, assembled and analysed at different levels. This study provides novel insights into the chloroplast genome evolutionary dynamics in the poorly studied Genistoid clade. Our results revealed highly conserved structure and gene content among the five Lupinus species with the exception of the rps16 gene, which is very likely pseudogenized in the different lupine species investigated. Detailed surveys of mitochondrion, nuclear and chloroplast genomes available to date revealed that rps16 gene is absent from all plant mitochondria, strongly suggesting that this gene was functionally replaced by the nuclear rps16 gene since the divergence of plants. Compared with the mitochondrion, the chloroplast rps16 gene is still present in many plants but has lost its functionality many times independently. Analysis of the evolution rate of functional rps16 genes present in both the nuclear and chloroplast genomes of some representative angiosperm species revealed that these genes are both under purifying selection, whereas a relaxed selective constraint was expected for the chloroplast copy. Comparative analyses of lupine plastomes also enabled identification of two hypervariable regions: psaA-ycf4 (11.5 kb) and ycf1-rps15 (6.5 kb). We demonstrate that these regions, which contain a high number of potentially informative sites and the highest number of SSRs, were highly consistent with, and reinforced the support for, previous phylogenies. The analyses of the short repeated sequences present in Lupinus plastomes allowed us to identify different types of chloroplast markers that could be very useful, low cost and easy to use for studying genetic diversity and evolutionary history of lupines or Genistoids.
Supplementary Material
Acknowledgments
The authors thank all institutions for kindly providing seed samples: Institut National d’Agronomie, Alger, Algérie; Institut National de la Recherche Agronomique, Dijon, France; Western Australian Department of Agriculture, Baron-Hay Court, South Perth, Western Australia; EMBRAPA-CENARGEN, Brasilia, Brasil; United States Department of Agriculture ARS, Washington DC, USA; University of Edmonton, Alberta, Canada.
The authors also thank M.T. Schifino-Wittman (Federal University of South Rio Grande, Brasil); D. Jones (University of Florida, USA); M.T. Misset (Université Rennes 1, France); R. Pasquet (IRD, Montpellier, France); J. Pascual (previously IMIA, Spain); M. Sahnoune (Université de Bejaia, Algeria). The authors greatly thank Thierry Fontaine-Breton and Fouad Nassur for taking care of the plant material. We also thank Christina Richards (Univ. of Florida, USA) and Jeremy Timmis (Univ. of Adelaide, Australia) for critically reading the manuscript as well as the two anonymous reviewers for their careful evaluation of our manuscript and their helpful comments and suggestions.
Conflict of interest
None declared.
Accession numbers
KX787895, KX787896, KX787897, KX787898, KX787899, KX787900, KX787901, KX787902, KX787903, KX787904, KX787905, KX787906, KX787907, KX787908, KX787909, KX787910, KX147685, KX147686, KX147687, KX147688, KX147689, KX147690, KX147691, KX147692, KX147693, KX147694, KX147695, KX147696, KX147697, KX147698, KX147699, KX147700, KX147701, KX147702, KX147703, KX147704, KX147705, KX147706, KX147707, KX147708, KX147709, KX147710, KX147711, KX147712, KX147713, KX147714, KX147715, KX147716, KX147717, KX147718, KX147719, KX147720, KX147721, KX147722, KX147723, KX147724, KX147725, KX147726, KX147727, KX147728, KX147729, KX147730, KX147731, KX147732, KX147733, KX147734, KX147735, KX147736, KX147737, KX147738, KX147739, KX147740, KX147741, KX147742, KX147743, KX147744, KX147745, KX147746, KX147747, KX147748, KX147749, KX147750, KX147751, KX147752, KX147753, KU726826, KU726827, KU726828, KU726829
Supplementary data
Supplementary Figs. S1–S7 and Tables S1–S13 are available at DNARES Online.
Funding
Jean Keller was supported by a doctoral research grant from the University of Rennes 1 - French Ministry of Higher Education and Research. Mathieu Rousseau-Gueutin was supported by the European Union Seventh Framework Programme (FP7-CIG-2013-2017; Grant no. 333709). This work benefited from the International Associated Laboratory “Ecological Genomics of Polyploidy” supported by CNRS (INEE, UMR CNRS 6553 Ecobio), University of Rennes 1, Iowa State University (Ames, USA).
References
- 1. Cronk Q., Ojeda I., Pennington R.T.. 2006, Legume comparative genomics: progress in phylogenetics and phylogenomics. Curr. Opin. Plant Biol., 9, 99–103. [DOI] [PubMed] [Google Scholar]
- 2. Lewis G.P. 2005, Legumes of the World. Royal Botanic Gardens, Kew. [Google Scholar]
- 3. LPWG. 2013, Towards a new classification system for legumes: progress report from the 6th International Legume Conference. South Afr. J. Bot., 89, 3–9. [Google Scholar]
- 4. Doyle J.J., Luckow M.. 2003, The rest of the iceberg. Legume diversity and evolution in a phylogenetic context. Plant Physiol., 131, 900–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Young N.D., Debellé F., Oldroyd G.E.D., et al. 2011, The Medicago genome provides insight into the evolution of rhizobial symbioses. Nature, 480, 520–524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Graham P.H., Vance C.P.. 2003, Legumes: importance and constraints to greater use. Plant Physiol., 131, 872–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Cai Z., Guisinger M., Kim H.-G., et al. 2008, Extensive reorganization of the plastid genome of Trifolium subterraneum (Fabaceae) is associated with numerous repeated sequences and novel DNA insertions. J. Mol. Evol., 67, 696–704. [DOI] [PubMed] [Google Scholar]
- 8. Doyle J.J., Doyle J.L., Palmer J.D.. 1995, Multiple independent losses of two genes and one intron from legume chloroplast genomes. Syst. Bot., 20, 272. [Google Scholar]
- 9. Guisinger M.M., Kuehl J.V., Boore J.L., Jansen R.K.. 2011, Extreme reconfiguration of plastid genomes in the angiosperm family Geraniaceae: rearrangements, repeats, and codon usage. Mol. Biol. Evol., 28, 583–600. [DOI] [PubMed] [Google Scholar]
- 10. Haberle R.C., Fourcade H.M., Boore J.L., Jansen R.K.. 2008, Extensive rearrangements in the chloroplast genome of Trachelium caeruleum are associated with repeats and tRNA genes. J. Mol. Evol., 66, 350–61. [DOI] [PubMed] [Google Scholar]
- 11. Jansen R.K., Ruhlman T.A.. 2012, Plastid genomes of seed plants In: Bock R., Knoop V. (eds.), Genomics of Chloroplasts and Mitochondria. Springer, Netherlands, Dordrecht, pp. 103–26. [Google Scholar]
- 12. Magee A.M., Aspinall S., Rice D.W., et al. 2010, Localized hypermutation and associated gene losses in legume chloroplast genomes. Genome Res., 20, 1700–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Weng M.-L., Blazier J.C., Govindu M., Jansen R. K.. 2014, Reconstruction of the ancestral plastid genome in Geraniaceae reveals a correlation between genome rearrangements, repeats, and nucleotide substitution rates. Mol. Biol. Evol., 31, 645–59. [DOI] [PubMed] [Google Scholar]
- 14. Dugas D.V., Hernandez D., Koenen E.J.M., et al. 2015, Mimosoid legume plastome evolution: IR expansion, tandem repeat expansions, and accelerated rate of evolution in clpP. Sci. Rep., 5, 16958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Martin G.E., Rousseau-Gueutin M., Cordonnier S., et al. 2014, The first complete chloroplast genome of the Genistoid legume Lupinus luteus: evidence for a novel major lineage-specific rearrangement and new insights regarding plastome evolution in the legume family. Ann. Bot., 113, 1197–210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Schwarz E.N., Ruhlman T.A., Sabir J.S.M., et al. 2015, Plastid genome sequences of legumes reveal parallel inversions and multiple losses of rps16 in papilionoids: parallel inversions and rps16 losses in legumes. J. Syst. Evol., 53, 458–68. [Google Scholar]
- 17. Sherman-Broyles S., Bombarely A., Grimwood J., Schmutz J., Doyle J.. 2014, Complete plastome sequences from Glycine syndetika and six additional perennial wild relatives of soybean. G3amp58 GenesGenomesGenetics, 4, 2023–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Williams A.V., Boykin L.M., Howell K.A., Nevill P.G., Small I.. 2015, The complete sequence of the Acacia ligulata chloroplast genome reveals a highly divergent clpP1 gene. C.-H. Kuo (ed.). PLOS ONE, 10, e0125768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Palmer J.D., Nugent J.M., Herbon L.A.. 1987, Unusual structure of geranium chloroplast DNA: a triple-sized inverted repeat, extensive gene duplications, multiple inversions, and two repeat families. Proc. Natl. Acad. Sci. U. S. A., 84, 769–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Doyle J.J., Doyle J.L., Ballenger J.A., Palmer J.D.. 1996, The distribution and phylogenetic significance of a 50-kb chloroplast DNA inversion in the flowering plant family Leguminosae. Mol. Phylogenet. Evol., 5, 429–438. [DOI] [PubMed] [Google Scholar]
- 21. Jansen R.K., Wojciechowski M.F., Sanniyasi E., Lee S.-B., Daniell H.. 2008, Complete plastid genome sequence of the chickpea (Cicer arietinum) and the phylogenetic distribution of rps12 and clpP intron losses among legumes (Leguminosae). Mol. Phylogenet. Evol., 48, 1204–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Wojciechowski M.F., Lavin M., Sanderson M.J.. 2004, A phylogeny of legumes (Leguminosae) based on analysis of the plastid matK gene resolves many well-supported subclades within the family. Am. J. Bot., 91, 1846–1862. [DOI] [PubMed] [Google Scholar]
- 23. Bruneau A., Doyle J.J., Palmer J.D.. 1990, A chloroplast DNA inversion as a subtribal character in the phaseoleae (Leguminosae). Syst. Bot., 15, 378. [Google Scholar]
- 24. Guo X., Castillo-Ramírez S., González V., et al. 2007, Rapid evolutionary change of common bean (Phaseolus vulgaris L.) plastome, and the genomic diversification of legume chloroplasts. BMC Genom., 8, 228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Tangphatsornruang S., Sangsrakru D., Chanprasert J., et al. 2010, The chloroplast genome sequence of mungbean (Vigna radiata) determined by high-throughput pyrosequencing: structural organization and phylogenetic relationships. DNA Res., 17, 11–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Kazakoff S.H., Imelfort M., Edwards D., et al. 2012, Capturing the biofuel wellhead and powerhouse: the chloroplast and mitochondrial genomes of the leguminous feedstock tree Pongamia pinnata. J.H. Badger (ed.). PLoS ONE, 7, e51687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Timmis J.N., Ayliffe M.A., Huang C.Y., Martin W.. 2004, Endosymbiotic gene transfer: organelle genomes forge eukaryotic chromosomes. Nat. Rev. Genet., 5, 123–35. [DOI] [PubMed] [Google Scholar]
- 28. Ueda M., Nishikawa T., Fujimoto M., et al. 2008, Substitution of the gene for chloroplast RPS16 was assisted by generation of a dual targeting signal. Mol. Biol. Evol., 25, 1566–75. [DOI] [PubMed] [Google Scholar]
- 29. Fleischmann T.T., Scharff L.B., Alkatib S., Hasdorf S., Schöttler M. A., Bock R.. 2011, Nonessential plastid-encoded ribosomal proteins in tobacco: a developmental role for plastid translation and implications for reductive genome evolution. Plant Cell, 23, 3137–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Kode V., Mudd E.A., Iamtham S., Day A.. 2005, The tobacco plastid accD gene is essential and is required for leaf development. Plant J., 44, 237–44. [DOI] [PubMed] [Google Scholar]
- 31. Shikanai T., Shimizu K., Ueda K., Nishimura Y., Kuroiwa T., Hashimoto T.. 2001, The chloroplast clpP gene, encoding a proteolytic subunit of ATP-dependent protease, is indispensable for chloroplast development in tobacco. Plant Cell Physiol., 42, 264–73. [DOI] [PubMed] [Google Scholar]
- 32. Jansen R.K., Cai Z., Raubeson L.A., et al. 2007, Analysis of 81 genes from 64 plastid genomes resolves relationships in angiosperms and identifies genome-scale evolutionary patterns. Proc. Natl. Acad. Sci. U. S. A., 104, 19369–19374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Cardoso D., de Queiroz L.P., Pennington R.T., et al. 2012, Revisiting the phylogeny of papilionoid legumes: new insights from comprehensively sampled early-branching lineages. Am. J. Bot., 99, 1991–2013. [DOI] [PubMed] [Google Scholar]
- 34. Sveinsson S., Cronk Q.. 2014, Evolutionary origin of highly repetitive plastid genomes within the clover genus (Trifolium). BMC Evol. Biol., 14, 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Gepts P., Beavis W.D., Brummer E.C., et al. 2005, Legumes as a model plant family. Genomics for food and feed report of the cross-legume advances through genomics conference. Plant Physiol., 137, 1228–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Gladstones J.S., Atkins C.A., Hamblin J. (eds.). 1998, Lupins as Crop Plants: Biology, Production, and Utilization. CAB International, Wallingford, Oxon, UK; New York, NY, USA. [Google Scholar]
- 37. Cabello-Hurtado F., Keller J., Ley J., Sanchez-Lucas R., Jorrín-Novo J.V., Aïnouche A.. 2016, Proteomics for exploiting diversity of lupin seed storage proteins and their use as nutraceuticals for health and welfare. J. Proteomics, 143, 57–68. [DOI] [PubMed] [Google Scholar]
- 38. Yang H., Tao Y., Zheng Z., et al. 2013, Draft genome sequence, and a sequence-defined genetic linkage map of the legume crop species Lupinus angustifolius. X.L. Cui, (ed.). PLoS ONE, 8, e64799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Ainouche A.-K., Bayer R.J.. 1999, Phylogenetic relationships in Lupinus (Fabaceae: Papilionoideae) based on internal transcribed spacer sequences (ITS) of nuclear ribosomal DNA. Am. J. Bot., 86, 590–607. [PubMed] [Google Scholar]
- 40. Drummond C.S., Eastwood R.J., Miotto S.T.S., Hughes C.E.. 2012, Multiple continental radiations and correlates of diversification in Lupinus (Leguminosae): testing for key innovation with incomplete taxon sampling. Syst. Biol., 61, 443–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Eastwood R.J., Drummond C.S., Schifino-Wittmann M.T., Hughes. 2008, Lupins for health and wealth: 12th International Lupin Conference, Fremantle, Western Australia, 14–18 September 2008. In: J.A., Palta, J.D., Berger, and International Lupin Association (eds.). International Lupin Association, Canterbury, New Zealand.
- 42. Hughes C., Eastwood R.. 2006, Island radiation on a continental scale: exceptional rates of plant diversification after uplift of the Andes. Proc. Natl. Acad. Sci. U. S. A., 103, 10334–10339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Käss E., Wink M.. 1997, Molecular phylogeny and phylogeography of Lupinus (Leguminosae) inferred from nucleotide sequences of therbcL gene and ITS 1 + 2 regions of rDNA. Plant Syst. Evol., 208, 139–167. [Google Scholar]
- 44. Mahé F., Markova D., Pasquet R., Misset M.-T., Aïnouche A.. 2011, Isolation, phylogeny and evolution of the SymRK gene in the legume genus Lupinus L. Mol. Phylogenet. Evol., 60, 49–61. [DOI] [PubMed] [Google Scholar]
- 45. Mahé F., Pascual H., Coriton O., et al. 2011, New data and phylogenetic placement of the enigmatic Old World lupin: Lupinus mariae-josephi H. Pascual. Genet. Resour. Crop Evol., 58, 101–14. [Google Scholar]
- 46. Aïnouche A., Bayer R.J., Misset M.-T.. 2004, Molecular phylogeny, diversification and character evolution in Lupinus (Fabaceae) with special attention to Mediterranean and African lupines. Plant Syst. Evol., 246. [Google Scholar]
- 47. Coissac E., Hollingsworth P.M., Lavergne S., Taberlet P.. 2016, From barcodes to genomes: extending the concept of DNA barcoding. Mol. Ecol, 25, 1423–8. [DOI] [PubMed] [Google Scholar]
- 48. Langmead B., Salzberg S.L.. 2012, Fast gapped-read alignment with Bowtie 2. Nat. Methods, 9, 357–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Wyman S.K., Jansen R.K., Boore J.L.. 2004, Automatic annotation of organellar genomes with DOGMA. Bioinformatics, 20, 3252–5. [DOI] [PubMed] [Google Scholar]
- 50. Krzywinski M., Schein J., Birol I., et al. 2009, Circos: an information aesthetic for comparative genomics. Genome Res., 19, 1639–1645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Pierleoni A., Martelli P.L., Fariselli P., Casadio R.. 2007, BaCelLo: a Balanced subCellular Localization predictor. Protoc. Exch, 22, 408–16. [DOI] [PubMed] [Google Scholar]
- 52. Boden M., Hawkins J.. 2005, Prediction of subcellular localization using sequence-biased recurrent networks. Bioinformatics, 21, 2279–86. [DOI] [PubMed] [Google Scholar]
- 53. Emanuelsson O., Nielsen H., Brunak S., von Heijne G.. 2000, Predicting subcellular localization of proteins based on their N-terminal amino acid sequence. J. Mol. Biol., 300, 1005–16. [DOI] [PubMed] [Google Scholar]
- 54. Blum T., Briesemeister S., Kohlbacher O.. 2009, MultiLoc2: integrating phylogeny and Gene Ontology terms improves subcellular protein localization prediction. BMC Bioinf., 10, 274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Small I., Peeters N., Legeai F., Lurin C.. 2004, Predotar: a tool for rapidly screening proteomes forN-terminal targeting sequences. Proteomics, 4, 1581–90. [DOI] [PubMed] [Google Scholar]
- 56. Michel F., Ferat J.L.. 1995, Structure and activities of group II introns. Annu. Rev. Biochem., 64, 435–61. [DOI] [PubMed] [Google Scholar]
- 57. Michel F., Kazuhiko U., Haruo O.. 1989, Comparative and functional anatomy of group II catalytic introns—a review. Gene, 82, 5–30. [DOI] [PubMed] [Google Scholar]
- 58. Lehmann K., Schmidt U.. 2003, Group II introns: structure and catalytic versatility of large natural ribozymes. Crit. Rev. Biochem. Mol. Biol., 38, 249–303. [DOI] [PubMed] [Google Scholar]
- 59. Roy S., Ueda M., Kadowaki K., Tsutsumi N.. 2010, Different status of the gene for ribosomal protein S16 in the chloroplast genome during evolution of the genus Arabidopsis and closely related species. Genes Genet. Syst., 85, 319–26. [DOI] [PubMed] [Google Scholar]
- 60. Kearse M., Moir R., Wilson A., et al. 2012, Geneious basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics, 28, 1647–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Yang Z. 2007, PAML 4: phylogenetic analysis by maximum likelihood. Mol. Biol. Evol., 24, 1586–91. [DOI] [PubMed] [Google Scholar]
- 62. Paradis E., Claude J., Strimmer K.. 2004, APE: analyses of phylogenetics and evolution in R language. Bioinformatics, 20, 289–90. [DOI] [PubMed] [Google Scholar]
- 63. Kimura M. 1980, A simple method for estimating evolutionary rates of base substitutions through comparative studies of nucleotide sequences. J. Mol. Evol., 16, 111–20. [DOI] [PubMed] [Google Scholar]
- 64. Yang Z., Nielsen R.. 2000, Estimating synonymous and nonsynonymous substitution rates under realistic evolutionary models. Mol. Biol. Evol., 17, 32–43. [DOI] [PubMed] [Google Scholar]
- 65. Kurtz S., Choudhuri J.V., Ohlebusch E., Schleiermacher C., Stoye J., Giegerich R.. 2001, REPuter: the manifold applications of repeat analysis on a genomic scale. Nucleic Acids Res., 29, 4633–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Saski C., Lee S.-B., Daniell H., et al. 2005, Complete chloroplast genome sequence of glycine max and comparative analyses with other legume genomes. Plant Mol. Biol., 59, 309–22. [DOI] [PubMed] [Google Scholar]
- 67. Mayer C. 2007, October, Phobos: Highly Accurate Search for Perfect and Imperfect Tandem Repeats in Complete Genomes by Christoph Mayer.
- 68. Rozen S., Skaletsky H.. 2000, Primer3 on the WWW for general users and for biologist programmers. Methods Mol. Biol. Clifton NJ, 132, 365–86. [DOI] [PubMed] [Google Scholar]
- 69. Katoh K., Misawa K., Kuma K., Miyata T.. 2002, MAFFT: a novel method for rapid multiple sequence alignment based on fast Fourier transform. Nucleic Acids Res., 30, 3059–3066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Felsenstein J. 1985, Confidence limits on phylogenies: an approach using the bootstrap. Evolution, 39, 783. [DOI] [PubMed] [Google Scholar]
- 71. Posada D. 2008, jModelTest: phylogenetic model averaging. Mol. Biol. Evol., 25, 1253–6. [DOI] [PubMed] [Google Scholar]
- 72. Tamura K., Stecher G., Peterson D., Filipski A., Kumar S.. 2013, MEGA6: molecular evolutionary genetics analysis version 6.0. Mol. Biol. Evol., 30, 2725–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Finn R.D., Bateman A., Clements J., et al. 2014, Pfam: the protein families database. Nucleic Acids Res., 42, D222–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Yoon H.S., Hackett J.D., Ciniglia C., Pinto G., Bhattacharya D.. 2004, A molecular timeline for the origin of photosynthetic eukaryotes. Mol. Biol. Evol., 21, 809–18. [DOI] [PubMed] [Google Scholar]
- 75. Allen J.F. 2003, Why chloroplasts and mitochondria contain genomes. Comp. Funct. Genomics, 4, 31–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Allen J.F., Puthiyaveetil S., Ström J., Allen C.A.. 2005, Energy transduction anchors genes in organelles: Problems and paradigms. BioEssays, 27, 426–35. [DOI] [PubMed] [Google Scholar]
- 77. Allen J.F. 2015, Why chloroplasts and mitochondria retain their own genomes and genetic systems: colocation for redox regulation of gene expression. Proc. Natl. Acad. Sci. U. S. A., 112, 10231–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Pfannschmidt T., Nilsson A., Tullberg A., Link G., Allen J.F.. 1999, Direct transcriptional control of the chloroplast genes psbA and psaAB adjusts photosynthesis to light energy distribution in plants. IUBMB Life, 48, 271–6. [DOI] [PubMed] [Google Scholar]
- 79. Maier U.-G., Zauner S., Woehle C., et al. 2013, Massively convergent evolution for ribosomal protein gene content in plastid and mitochondrial genomes. Genome Biol. Evol., 5, 2318–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Kikuchi S., Bédard J., Hirano M., et al. 2013, Uncovering the protein translocon at the chloroplast inner envelope membrane. Science, 339, 571–574. [DOI] [PubMed] [Google Scholar]
- 81. Dong W., Xu C., Li C., et al. 2015, ycf1, the most promising plastid DNA barcode of land plants. Sci. Rep., 5, 8348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Dong W., Liu J., Yu J., Wang L., Zhou S.. 2012, Highly variable chloroplast markers for evaluating plant phylogeny at low taxonomic levels and for DNA barcoding. A. Moustafa, (ed.). PLoS ONE, 7, e35071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Swofford D.L. 2001, PAUP*: Phylogenetic Analysis Using Parsimony (and other methods) 4.0.b5.
- 84. Johnson L.A., Soltis D.E.. 1998, Assessing congruence: empirical examples from molecular data In: Soltis D.E., Soltis P.S., Doyle J.J., (eds.), Molecular Systematics of Plants II. Springer, US, pp. 297–348. [Google Scholar]
- 85. Drummond C.S. 2008, Diversification of Lupinus (Leguminosae) in the western New World: derived evolution of perennial life history and colonization of montane habitats. Mol. Phylogenet. Evol., 48, 408–21. [DOI] [PubMed] [Google Scholar]
- 86. Cosner M.E., Jansen R.K., Palmer J.D., Downie S.R.. 1997, The highly rearranged chloroplast genome of Trachelium caeruleum (Campanulaceae): multiple inversions, inverted repeat expansion and contraction, transposition, insertions/deletions, and several repeat families. Curr. Genet., 31, 419–429. [DOI] [PubMed] [Google Scholar]
- 87. Milligan B.G., Hampton J.N., Palmer J.D.. 1989, Dispersed repeats and structural reorganization in subclover chloroplast DNA. Mol. Biol. Evol., 6, 355–68. [DOI] [PubMed] [Google Scholar]
- 88. Ogihara Y., Terachi T., Sasakuma T.. 1988, Intramolecular recombination of chloroplast genome mediated by short direct-repeat sequences in wheat species. Proc. Natl. Acad. Sci. U. S. A., 85, 8573–8577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Akkaya M.S., Bhagwat A.A., Cregan P.B.. 1992, Length polymorphisms of simple sequence repeat DNA in soybean. Genetics, 132, 1131–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Desiderio F., Bitocchi E., Bellucci E., et al. 2013, Chloroplast microsatellite diversity in Phaseolus vulgaris. Front. Plant Sci., 3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Pan L., Li Y., Guo R., Wu H., Hu Z., Chen C.. 2014, Development of 12 chloroplast microsatellite markers in Vigna unguiculata (Fabaceae) and amplification in Phaseolus vulgaris. Appl. Plant Sci., 2, 1300075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Powell W., Morgante M., Andre C., et al. 1995, Hypervariable microsatellites provide a general source of polymorphic DNA markers for the chloroplast genome. Curr. Biol., 5, 1023–1029. [DOI] [PubMed] [Google Scholar]
- 93. Provan J., Powell W., Hollingsworth P.M.. 2001, Chloroplast microsatellites: new tools for studies in plant ecology and evolution. Trends Ecol. Evol., 16, 142–147. [DOI] [PubMed] [Google Scholar]
- 94. Provan J., Russell J.R., Booth A., Powell W.. 1999, Polymorphic chloroplast simple sequence repeat primers for systematic and population studies in the genus Hordeum. Mol. Ecol., 8, 505–511. [DOI] [PubMed] [Google Scholar]
- 95. Saghai Maroof M.A., Biyashev R.M., Yang G.P., Zhang Q., Allard R.W.. 1994, Extraordinarily polymorphic microsatellite DNA in barley: species diversity, chromosomal locations, and population dynamics. Proc. Natl. Acad. Sci. U. S. A., 91, 5466–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96. Wu K.-S., Tanksley S.D.. 1993, Abundance, polymorphism and genetic mapping of microsatellites in rice. Mol. Gen. Genet. MGG, 241, 225–235. [DOI] [PubMed] [Google Scholar]
- 97. Burki F. 2014, The eukaryotic tree of life from a global phylogenomic perspective. Cold Spring Harb. Perspect. Biol., 6, a016147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98. Leliaert F., Smith D.R., Moreau H., et al. 2012, Phylogeny and molecular evolution of the green algae. Crit. Rev. Plant Sci., 31, 1–46. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.