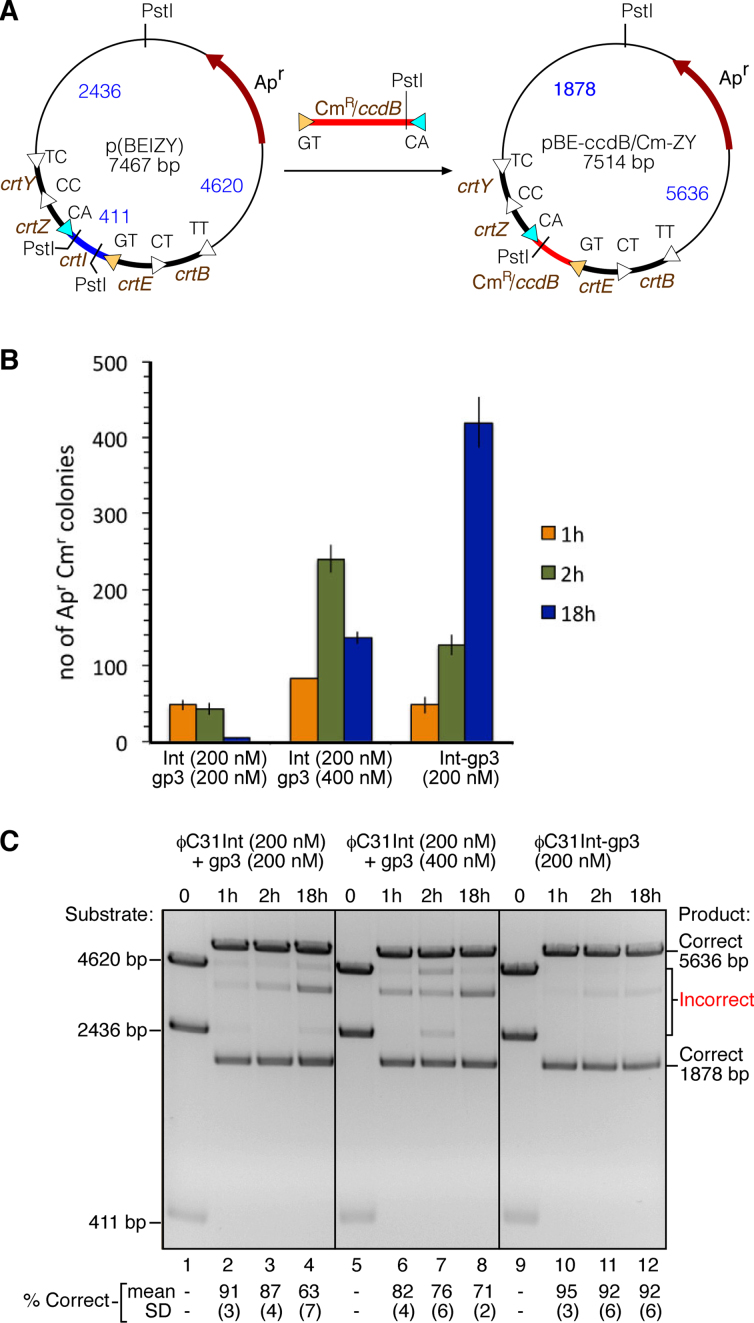
Figure 6.
Targeted editing of a gene array by an integrase–RDF fusion recombinase. (A) integrase–RDF fusion protein-mediated replacement of the crtI gene cassette in plasmid p(BEIZY) with a different cassette (CmR/ccdB). p(BEIZY) contains an array of genes crtB, crtE, crtI, crtZ and crtY, each separated by ϕC31 attL sites differing in their central dinucleotide sequence (8). The linear PCR product (red, cartooned above the arrow) contains the CmR/ccdB gene cassette (conferring chloramphenicol resistance), flanked by ϕC31 attR sites (attR-GT and attR-CA; yellow and cyan triangles) with central dinucleotides matching the attL sites flanking crtI. The usefulness of CmR/ccdB cassette exchange is described in (8); the properties of the ccdB gene are not used in the experiments described here. (B) Cassette substitution reactions were carried out in vitro with integrase and gp3 or Int-gp3 fusion recombinase, for varying times, as indicated on the figure. Reaction products were used to transform E. coli cells, transformants were selected on plates containing ampicillin and chloramphenicol, and colonies were counted. Each bar in the figure represents mean ± standard deviation from three experiments. (C) Plasmid DNA was recovered from the pooled E. coli colonies (as in B) and digested with PstI. The digest products were separated by agarose gel electrophoresis, and the proportion (%) of correctly assembled plasmids was estimated by quantitation of the bands. Quantitation values and standard deviation from triplicate experiments are given below each lane.