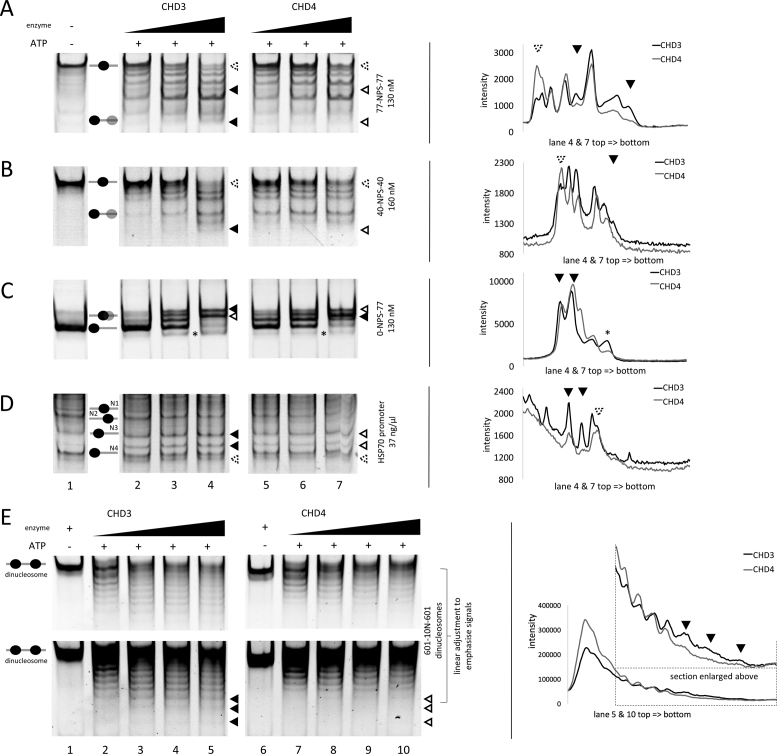
Figure 2.
Recombinant CHD3 and CHD4 exhibit distinct, sequence-specific nucleosome positioning behaviour. Recombinantly purified human CHD3/4 were titrated in increasing concentrations (A/C/E: 25, 50, 75 and 100 nM; B/D: 50, 100 and 200 nM) to the indicated mono- and dinucleosomal templates containing different configurations of linker DNA (see also Materials and Methods). The reactions were started by adding ATP. Nucleosomes in the absence or presence of enzyme (without ATP) served as reference. To visualize the nucleosome movements, the reactions were loaded on PAA gels (ethidium bromide stain). Positions of mono- and dinucleosomes are indicated by ovals, according to (68). Filled triangles represent more intense bands, empty triangles less intense bands or no signal, comparing CHD3 and CHD4 remodeling patterns. Triangles with a dashed rim were added for better orientation in the intensity profiles (see below). Black asterisks indicate nucleosomes, which were probably pushed over the edge of the DNA strand. On the right side of each remodeling gel are intensity profiles of the indicated gel lanes, based on Multi Gauge software analysis. The triangles indicate the positions of the bands highlighted by the triangles in the respective lanes of the corresponding gel picture. All lanes shown for one remodeling template are from one gel. Number or replicates: (A) ≥3; (B) 3; (C) ≥3; (D) 3; (E) ≥3.