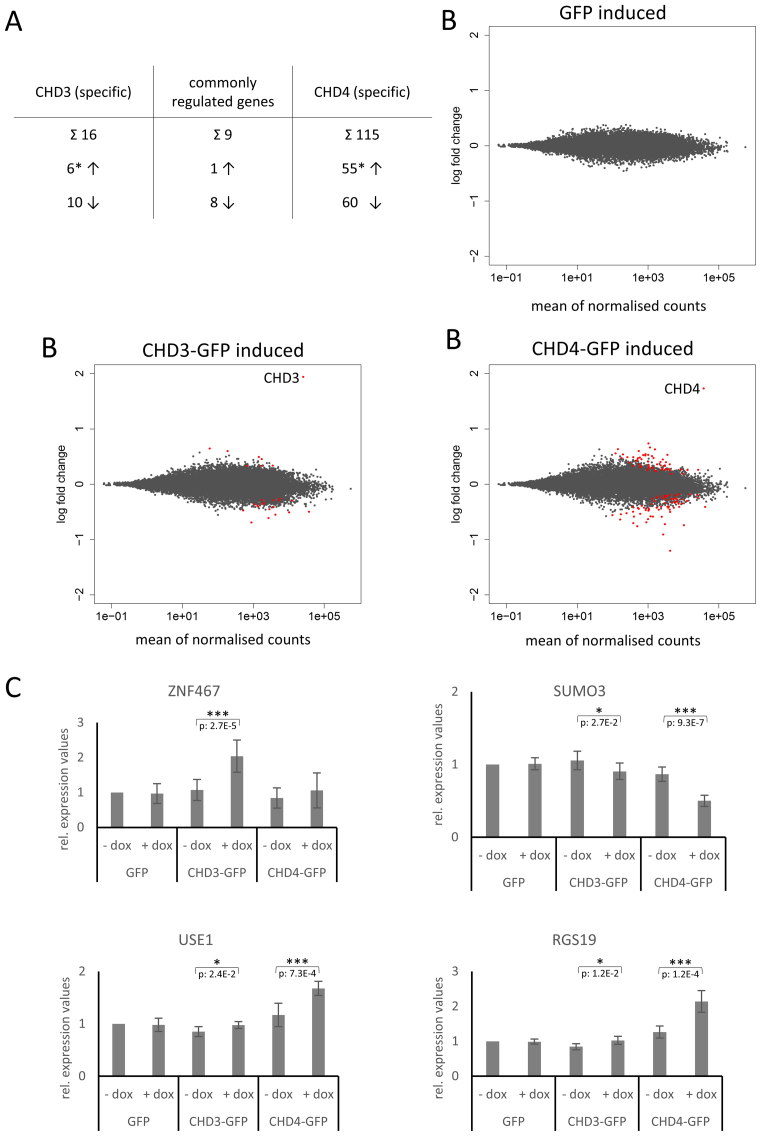
Figure 5.
CHD3/4 containing NuRD complexes regulate distinct genes in living cells. Stably transfected Flp-In™ T-Rex™ 293 cells were incubated in the absence (non-induced) or presence (induced) of doxycycline for 24 h. Five independent sets of total RNA were prepared. Two sets of total RNA were subjected to Illumina Sequencing (A and B): differential gene expression was analysed using the DESeq2 R package (59). Samples without doxycycline induction were considered as ground state and differential expression was determined by a false discovery rate below 0.1. (A) The table presents the total number of specifically and commonly up- (↑) and downregulated (↓) genes upon CHD3/4 expression (* the number of specifically upregulated genes does not include the upregulated CHD3/4 genes due to doxycycline treatment). (B) MA-plots illustrate log2 fold changes (y-axis) for each gene in comparison to the average count (x-axis). The red points indicate differentially expressed genes determined by DESeq2 analysis (FDR < 0.1), including the doxycycline induced CHD3/4 proteins. (C) The other three sets of total RNA were reversely transcribed into cDNA for qPCR experiments: Data were collected by the Rotor-Gene 3000 system, analyzed using the comparative analysis software module and presented as relative expression values, using Lamin A/C for normalization. The mean values and standard deviations are derived from three biological replicates (with at least two technical replicates for each biological replicate). Relative expression values for induced (+dox) and non-induced (–dox) conditions were tested for significance using a two tailed, homoscedastic t-test in Microsoft Excel. P-values are given for sets with significant changes (*P < 0.05, **P < 0.01, ***P < 0.001). Number of replicates: (A/B) 2 (see above); (C) 3 (see above) → in total: five independent total RNA preparations (see above). The raw data for the RNA-seq analysis are deposited in the GEO database (accession number: GSE93543).