Figure 6.
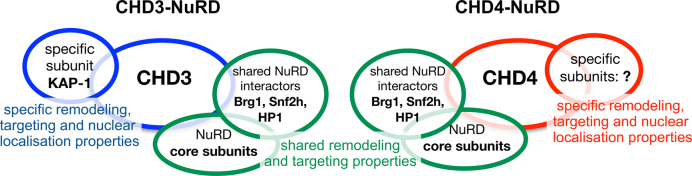
CHD3 and CHD4 form distinct NuRD complexes with different yet overlapping functionality. Our experiments propose that CHD3 and CHD4 form isoform-specific NuRD complexes in living cells, harboring one single copy of the respective remodeling enzyme. Beside well known ‘shared’ core subunits like HDAC1/2 or MTA2/3 proteins, we found Brg1, Snf2h and HP1 to be associated with both CHD3- and CHD4-containing NuRD complexes. DNA damage experiments, performed in this study, suggest that both CHD3- and CHD4-containing NuRD complexes exert a role in the DNA damage response. Nevertheless, an individual role of mouse CHD3 in DSB repair was proposed by the CHD3 specific binding protein KAP-1 (11,81). In addition to interaction partners that might act as functional NuRD regulators, we propose intrinsic remodeling properties as another possibility to influence the physiological role of NuRD complexes in vivo. Nucleosome remodeling assays with recombinant, single CHD3 and CHD4 proteins revealed that both enzymes move nucleosomes to distinct, sequence-specific positions, supporting the idea of CHD remodeling enzymes acting as chromatin specific organizers. In line with this, we observe mainly distinct nuclear localization patterns for CHD3- and CHD4-NuRD complexes in living cells, arguing for the existence of structurally and spatially separated complexes acting independently from each other in different genomic regions with a putative effect on gene activity. Indeed, our RNA-seq and qPCR experiments showed that CHD3 and CHD4 mainly regulate distinct genes. Taken together our data suggest that CHD3 and CHD4 form distinct NuRD complexes with different yet overlapping functionality (see also Results and Discussion).