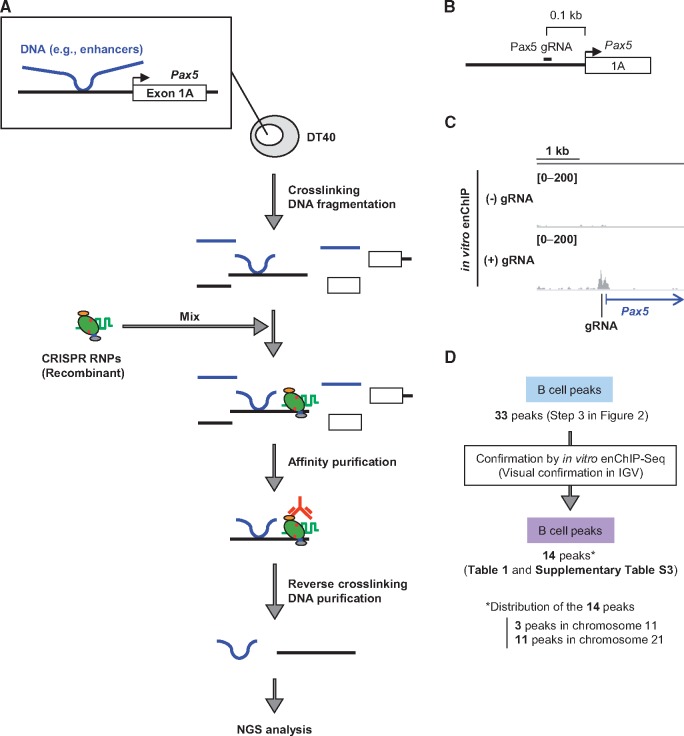
Figure 3.
In vitro enChIP-Seq for confirmation of the results of iChIP-Seq. (A) Intact DT40 cells were crosslinked with formaldehyde, and chromatin DNA was fragmented by sonication. Recombinant CRISPR ribonucleoproteins (RNPs), which consist of 3xFLAG-dCas9-Dock and gRNA targeting the Pax5 promoter region, were mixed with the fragmented chromatin DNA to capture the target region. After affinity purification with anti-FLAG antibody, reversal of crosslinking, and DNA purification, the DNA was subjected to NGS analysis. (B) Target position of Pax5 gRNA. (C) NGS peak images around the Pax5 promoter region. NGS data from in vitro enChIP-Seq were visualized in IGV. The vertical viewing range (y-axis shown as scale) was set at 0-200 based on the magnitude of the noise peaks. (D) Confirmation of the results of iChIP-Seq. The peak positions identified by iChIP-Seq were confirmed by in vitro enChIP-Seq in IGV.