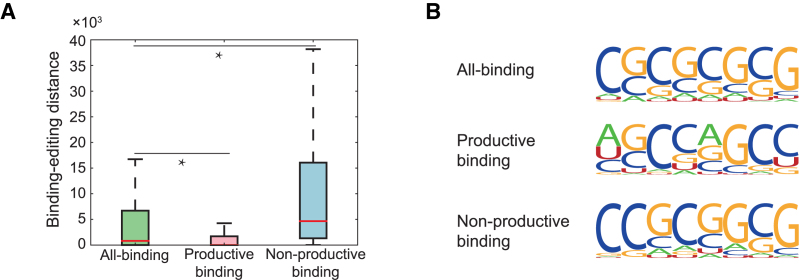
Figure 6.
The comparison results on three different DeBooster models, which were trained using all ADAR1 binding sites identified by CLIP-seq experiments, productive ADAR1 binding sites (i.e. triggering A-to-I editing), and non-productive ADAR1 binding sites (i.e. without triggering A-to-I editing), respectively. (A) The boxplot of the binding-editing distances, which were defined as the genomic distances between the new predicted ADAR1 binding sites and the closet editing sites, for three different DeBooster models. *P value < 0.001, Wilcoxon rank sum test. (B) The sequence motifs of the ADAR1 binding sites identified by three different DeBooster models. More details can be found in the main text.