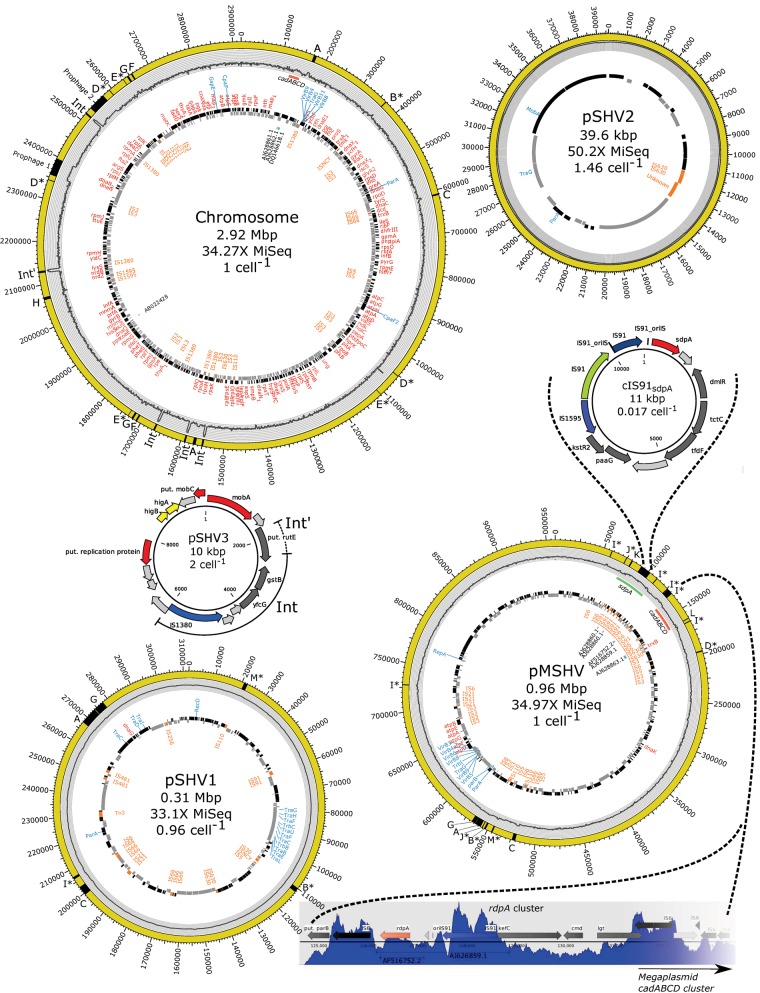
Fig. 1.
—The four major replicons of Sphingobium herbicidovorans MH (yellow circles: Chromosome, Megaplasmid pMSHV, pSHV1, and pSHV2) and minor circular elements (10-kb mobilizable plasmid pSHV3 and 11-kb IS91-mediated circular element cIS91sdpA) determined from sequencing of exonuclease treated DNA (mobilome data). Relative sizes of circles do not represent their actual scale. Black and gray tiles represent CDS features, as predicted by PROKKA, on the forward and reverse strand, respectively. Identified housekeeping genes are marked in red, plasmid-backbone genes are in blue, and IS elements are in orange on the inside of the CDS features. Previously studied genes and PCR clones are marked as their GenBank accession numbers in black. The sdpA (green) and cadABCD (red) gene clusters on major replicons are highlighted just under the mobilome histogram. The histogram on the gray background shows the mapping of paired-end reads from the exonuclease-treated mobilome sequencing. The mobilome coverage was log-transformed and histograms for all replicons have the same scale for direct comparison (e.g., the average mobilome coverage of the chromosome is much lower than that of pSHV2). Putative circular elements from the mobilome data are highlighted in black on the outer yellow diagram (A–M) and are shown in further detail in figure 4. The mobilizable plasmid pSHV3 has been partially integrated five separate times into the chromosome (Int) of which one integration is longer than the other four and contains the full rutE ORF, encoding a putative nitroreductase-like family protein, besides the yfcG and gstB genes encoding putative a GSH-dependent disulfide bond oxidoreductase and a putative glutathione S-transferase. For pSHV3, cIS91sdpA, and rdpA cluster arrows indicate hypothetical genes (light gray), miscellaneous genes (dark gray), toxin–antitoxin genes (yellow), and genes of special interest (other colors).