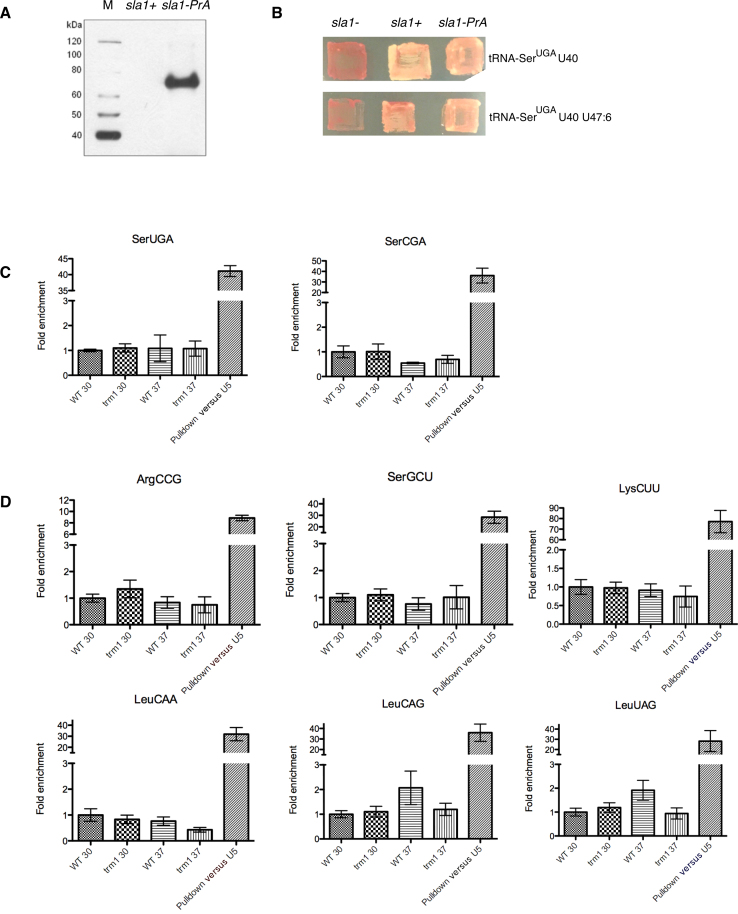
Figure 5.
Association of pre-tRNAs with Sla1p-PrA +/− G26 modification in Schizosaccharomyces pombe cells. Endogenous Sla1p-PrA/pre-tRNA complexes from trm1− and trm1+ cells grown at 30°C versus 37°C were immunoprecipitated and levels of candidate pre-tRNAs were quantitated by ‘TaqMan’ quantitative real-time PCR (qPCR). (A) Western blot confirming the expected size and expression of the Sla1-PrA fusion protein relative to the parent sla1+ strain. (B) Endogenous Sla1p containing an integrated Protein-A tag at the C-terminus (sla1-PrA) is comparably active to WT Sla1p as determined by a tRNA mediated suppression assay using two different defective suppressor tRNA alleles (8). (C) Analysis of fold pulldown levels of candidate misfolded pre-tRNAs (as measured by impaired charging and/or chemical probing, Figure 2) compared to an internal control, the geometric mean pulldown level of three pre-tRNAs not modified by Trm1p (pre-tRNA-IleUAU, pre-tRNA-TyrGUA and pre-tRNA-AlaCGC) in the same elution samples. As a reference for robust pulldown of La targets versus non-targets, fold pulldown of indicated pre-tRNA in elution samples versus total RNA samples (WT strain) is provided relative to the La non-target U5 snRNA (right hand side, histograms). (D): same as for (C) but for candidate pre-tRNAs identified as having decreased accumulation in sla1−/trm1− strains (see Figure 4).