Abstract
Background and Aims Polyploidy is arguably the single most important genetic mechanism in plant speciation and diversification. It has been repeatedly suggested that polyploids show higher vegetative reproduction than diploids (to by-pass low fertility after the polyploidization), but there are no rigorous tests of it.
Methods Data were analysed by phylogenetic regressions of clonal growth parameters, and vegetative reproduction in culture on the ploidy status of a large set of species (approx. 900) from the Central European Angiosperm flora. Further, correlated evolution of ploidy and clonal traits was examined to determine whether or not polyploidy precedes vegetative reproduction.
Key Results The analyses showed that polyploidy is strongly associated with vegetative reproduction, whereas diploids rely more on seed reproduction. The rate of polyploid speciation is strongly enhanced by the existence of vegetative reproduction (namely extensive lateral spread), whereas the converse is not true.
Conclusions These findings confirm the old hypothesis that polyploids can rely on vegetative reproduction which thus may save many incipient polyploids from extinction. A closer analysis also shows that the sequence of events begins with development of vegetative reproduction, which is then followed by polyploidy. Vegetative reproduction is thus likely to play an important role in polyploid speciation.
Keywords: polyploidy, vegetative reproduction, phylogenetic analysis, clonal traits, correlated evolution
INTRODUCTION
Polyploidization (i.e. the multiplication of the whole chromosome complement above the diploid state) is assumed to be one of the major mechanisms of plant speciation and evolution (Otto and Whitton, 2000; Husband et al., 2013). It has been shown that all extant seed plants have undergone one or more ancient polyploidization events (Jiao et al., 2011; Soltis et al., 2014) and there is cytological evidence for more recent whole-genome duplication(s) in many plant groups, particularly angiosperms and ferns (Wood et al., 2009; Husband et al., 2013). Establishment of a new polyploid lineage can happen through several different evolutionary pathways (Fowler and Levin, 1984; Felber, 1991, Ramsey and Schemske, 1998; Husband, 2004), but almost invariably neopolyploid derivatives face strongly reduced fitness, due either to mating with their more common diploid/lower polyploid ancestors (minority cytotype disadvantage; Levin, 1975) or to meiotic irregularities, namely the formation of multivalents in autopolyploids (Comai, 2005; Weiss-Schneeweiss et al., 2013). These processes probably counteract establishment of polyploid lineages and constrain the observed rates of polyploid speciation (Otto and Whitton, 2000; Meyers and Levin, 2006; Wood et al., 2009).
The underlying processes by which polyploids cope with reproductive disadvantages in early stages after their formation are still poorly understood (Soltis et al., 2010; Weiss-Schneeweiss et al., 2013). Polyploids often escape this disadvantage if they are self-compatible (Robertson et al., 2011), but it has long been hypothesized that another viable mechanism to escape the reduced sexual reproduction (i.e. via seeds formed through sexual means) is vegetative reproduction whereby genetically identical ramets are produced through clonal growth (von Wettstein, 1927; Gustafsson, 1948; Stebbins, 1957; see also Husband et al., 2013). Clonal reproduction enables newly formed polyploids to persist by forming sufficiently large populations in the absence of sexual reproduction. In facultatively sexual species, clonality may increase the degree of within-ploidy pollinator foraging and pollination, thus reducing the strength of minority cytotype disadvantage. In addition, the prevalence of vegetative propagation may allow neopolyploids to persist long enough for additional sexually compatible polyploid(s) to be formed within a population. In contrast to a large body of literature exploring the link between polyploidy and apomixis (e.g. Whitton et al., 2008), only a few studies addressed the association between karyological properties and the intensity of vegetative reproduction. While some comparisons at the intra-specific level failed to find any inter-ploidy differences (e.g. Keeler, 2004; Baldwin and Husband, 2013), others reported greater vegetative reproduction in (high) polyploids as compared with their diploid/lower polyploid counterparts (Eckert et al., 2003; Duchoslav and Staňková, 2015). The only attempt to understand the large-scale pattern was made nearly seven decades ago by Åke Gustafsson (Gustafsson, 1948). By comparing 36 genera in weed floras of Sweden, Switzerland and Canada, he concluded that ‘root-wandering’ perennials (i.e. species possessing an uninterrupted capacity for vegetative spreading) contain a markedly higher proportion of polyploids than annuals and biennials (i.e. species reproducing exclusively by seeds), and perennials incapable of vegetative dispersal (called ‘stationery perennials’).
Although the capability for vegetative propagation has become a textbook example of factors promoting polyploidy (Grant, 1981), there are several issues that do not stand closer scrutiny. First, Gustafsson’s (1948) analyses did not account for phylogenetic relationships. Although he examined relationships between vegetative reproduction and polyploidy also within individual plant groups (essentially applying phylogenetically independent contrasts informally), appropriate analysis of the overall pattern is still lacking. Secondly, assessment of the role of vegetative reproduction across large species sets is not as trivial as it may seem. There are no good data on the prevalence of vegetative (clonal) relative to sexual (seed) reproduction, because data on vegetative reproduction are difficult to collect and are available only for a small part of the flora (Klimešová and de Bello, 2009). In addition, it is not clear to what extent such data are informative on the actual role of vegetative relative to sexual reproduction. While several studies implied negative trade-off between both reproductive modes (Obeso, 2002; Boedeltje et al., 2008; Herben et al., 2012), trade-off relationships may be concealed by other sources of variation and need not contribute significantly to the observed pattern (Reznick et al., 2000).
Most importantly, however, correlation of polyploidy and vegetative reproduction, even when supported by good data, may arise due to a number of processes. Clonal plants may be predisposed to polyploid speciation because neopolyploids would be shielded from initial adverse fitness effects of genome duplication by vegetative reproduction and thus are more likely to overcome the minority disadvantage (see Husband et al., 2013). Alternatively, vegetative reproduction may be triggered by polyploidization through accompanying changes in cell morphology and/or metabolic and developmental rates (Otto and Whitton, 2000). It has been shown that the number of meristematic cells increases after genome duplication, which affects the dynamics of cell division (Asker and Jerling, 1992). The correlation between polyploidy and vegetative reproduction may also arise due to links of both processes to another factor, and polyploid-clonal would be expected without clonality directly interacting with polyploid establishment. Hence the key question in the study of the evolutionary role of vegetative reproduction and polyploidy in speciation is to understand the temporal sequence of both processes and thus assess support for the ‘pre-adaptation scenario’ vs. the ‘evolution of clonality’, or lack of relationship if the correlation is due to a third factor.
We believe that the time has come to reassess these, essentially almost century-old, questions in the light of newly collected data and new techniques. Namely, the increasing availability of phylogenetic information offers the possibility to explore trait correlations with proper account of species non-independence, and to model the evolution of a (potential) relationship between vegetative reproduction and polyploidy. Further, we are in possession of much better data on vegetative reproduction in large sets of species. First, the CLO-PLA database (database of clonal and bud bank traits of Central European flora; Klimešová and de Bello, 2009) provides data on a number of parameters of clonal growth, including bud bank size, lateral spread and persistence of spacer connections. Secondly, there is a unique data set on the actual role of vegetative and seed reproduction of a large number of species (Herben et al., 2012) that can be used to assess the relative importance of both reproductive modes. Both these data sets cover floras with very good knowledge of karyological variation of the component taxa, thus making meaningful comparisons possible.
We therefore examined relationships between ploidy level and intensity of vegetative reproduction in a phylogenetically diverse set of nearly 1000 angiosperm species from the central European flora. We used information on vegetative reproduction parameters from the CLO-PLA database (Klimešová and de Bello, 2009) and complemented it with data on the extent of seed and vegetative reproduction using long-term observation data (see also Herben et al., 2012, 2014). These data were linked to the ploidy level(s) of each species to identify the type and strength of the correlation between ploidy level and seed/vegetative reproduction, taking into account phylogenetic relatedness of the taxa. We compared these analyses with non-phylogenetic analyses to determine to what extent these correlations are due to shared evolutionary history. We also examined different macroevolutionary optima for traits of vegetative reproduction between diploids and polyploids using the Ohrenstein–Uhlenbeck (OU) model (Beaulieu et al., 2012). As the second step, we modelled correlated evolution of polyploidy and vegetative reproduction to identify whether existing data provide any information on the temporal sequence of both processes (i.e. whether polyploidy follows the appearance of vegetative reproduction or vice versa).
MATERIALS AND METHODS
Ploidy data
Data on ploidy levels were extracted from karyological indices of vascular plants of the Czech Republic and neighbouring countries, including the internal karyological database of plants of the Czech Republic (held at the Institute of Botany CAS, Průhonice), Karyological database of the ferns and flowering plants of Slovakia (Marhold et al., 2012), Chromosome number database of the Polish flora (Góralski et al., 2009 onwards) and the Documented chromosome number checklist of Austria – vascular plants (Dobeš and Vitek, 2000). The Index to plant chromosome numbers (Goldblatt and Johnson, 1979 onwards) and the Chromosome Counts Database (Rice et al., 2015) were also consulted in order to obtain global insight into the ploidy homogeneity/heterogeneity of analysed species, as they contain chromosomal data from different geographic regions that permit us to check whether a species is ploidy-uniform or variable. Aneuploid counts (typically differing only by one or two chromosomes from the nearest euploid number) were replaced by the nearest euploid number. The putative basic chromosome number (x) was determined for each species by comparing available somatic chromosome numbers of congeners and occasionally also of representatives of closely related genera. In general, x > 12 was considered as secondary (e.g. Goldblatt, 1980; Rice et al., 2015), although exceptions existed (see Supplementary Data Table S1). Two ploidy categories (further referred to as ploidy types) were distinguished: (1) diploids (i.e. diploid species with a primary basic chromosome number; 415 species) and (2) polyploids. The latter included both species harbouring a diploid number of somatic chromosomes but with a secondary basic chromosome number, most probably originating via (ancient) allopolyploidy (79 species), and species harbouring more than three basic chromosome sets in somatic cells (446 species). We did not distinguish between intra-specific (autopolyploidy) and inter-specific (allopolyploidy) polyploidization.
Although probabilistic methods to infer a putative base chromosome number (and thus ploidy level) are now available (e.g. the ChromEvol program of Glick and Mayrose, 2014), we preferred educated guesses to distinguish between the ploidy types. In our opinion, our data are not suitable for a meaningful analysis using probabilistic methods as (1) both auto- and allopolyploids (with often complex and poorly known evolutionary history) are included and (2) different types of transitions between chromosome numbers occurred in individual clades. Specifically, allopolyploids with high chromosome numbers and with all diploid progenitors extinct would be reconstructed as diploid using the ChromEvol program.
In ploidy-variable species, only ploidies previously reported from the Czech Republic or likely to occur there (based on records from neighbouring countries) were considered. All individuals of species known to have several cytotypes in the Czech Republic for which reproduction scores (see below) were available from the botanical garden were subjected to flow cytometric estimation of nuclear genome size (following the methodology detailed by Doležel et al., 2007). The resulting 2C-values were compared with data in the Plant DNA C-values Database (Bennett and Leitch, 2012) and, whenever possible, the ploidy level of the analysed sample was inferred. If this approach failed, the species was excluded unless one of the cytotypes was known to be considerably more common in the Czech Republic than the other(s). In case of doubt. the species was also excluded from the analysis. Only species with even ploidy levels were used in the analysis. We excluded odd-ploidy cytotypes because they constituted only a small minority of analysed samples and their sexual reproduction is hindered by a high level of sterility due to meiotic irregularities (Ramsey and Schemske, 1998). The final data set contained 940 Angiosperm species with a known ploidy level (see Table S1).
Data on phylogeny and species traits
Clonal growth data are taken from the database of clonal traits (CLO-PLA, version 3.3, J. Klimešová, unpubl.; see also Klimešová and de Bello, 2009). We transformed the data from the database to yield four traits (see Table 1; Supplementary Data Table S2). We used three traits: (1) lateral spreading distance as a measure of mean distance between mother and its clonal offspring; (2) number of clonal offspring as a measure of (potential) clonal multiplication; and (3) bud bank size as a measure of the potential of the plant to resprout after disturbance. While these traits are known to vary intraspecifically in response to environment, inter-specific differences from the database have been shown to contain a meaningful and robust signal (see, for example, Klimešová et al., 2016).
Table 1.
Traits used in the analyses
| Name | Units | Definition | No. of species with known ploidy for which data are available | No. of species with known ploidy and phylogeny for which data are available |
|---|---|---|---|---|
| Clonal traits | ||||
| Bud bank size | Count | No. of stem-derived buds per mother plant shoot in the soil and at the soil surface | 766 | 762 |
| No. of clonal offspring* | Count | No. of clonal offspring shoots per parent shoot per year, including offspring of small size. Small offspring are defined as those clonal offspring for which it took more years to attain size comparable with other clonal offspring of the plant; they usually resemble seedlings | 480 | 477 |
| Lateral spreading* | m | Lateral spreading distance of clonal growth organs | 480 | 477 |
| Reproduction scores | ||||
| Seed reproduction | Ordinal | Score (1–5) | 922 | 877 |
| Vegetative reproduction | Ordinal | Score (1–5) | 967 | 922 |
Trait is defined only for plants with the ability for clonal growth.
Phylogenetic data were taken primarily from Daphne (Durka and Michalski, 2012). Species missing from the original Daphne phylogeny (65 taxa) were added following Lososová et al. (2015). If no data were available for a given species, the species was excluded from the phylogenetic analysis. This approach yielded independent phylogenetic information for 895 species with known ploidy levels.
Data on seed and vegetative reproduction
Data on actual vegetative and seed reproduction were gathered from the collection of native plants of the Central European Flora in the Botanical Garden of the Faculty of Science, Charles University in Prague (http://www.bz-uk.cz; see also Herben et al., 2012). The collection houses about 1200 central European plant species, collected in the Czech Republic and Slovakia. Plants have been collected over an extensive period, beginning in the 1930s, although most of them were collected in 1960–2000. Each species has been kept under conditions that we assume to be as close as possible to their natural conditions within the garden. The habitats in the garden range in moisture from open, dry, sandy habitats and limestone, rocky habitats through mesic open habitats and shaded forest stands to moist (shaded and unshaded) places. Plants have been grown in open soil, with weeding carried out, including removal of individuals of the planted species, in order to keep stands of each species separate.
All plant species that have been growing in the garden for > 10 years were assigned five-degree scores for vegetative and seed reproduction for that period. The scores express the estimated frequency of thinning of given species on an approximately logarithmic scale. Species that multiply spontaneously were given a score of 3, 4 and 5 depending on the frequency of necessary thinning (one in several years, yearly and several times a year, respectively), species that do not multiply spontaneously, but can be multiplied by simple outdoor gardening techniques (splitting tussocks, planting cuttings, sowing seed, etc.) were given a score of 2 and species that do not multiply in the garden were given a score of 1 (for further details, see Herben et al., 2012). Seed and vegetative reproduction were scored separately using the same ordinal scale by one person and are further referred to as reproduction scores. In most cases, seedlings could be distinguished from vegetative offspring. However, in some plants with vigorous vegetative reproduction, assessment of seed reproduction was impossible due to potential seedlings being mixed with the vegetative progeny, and seed reproduction in these plants had to be treated as a missing value. Plants that are maintained in several habitats (or plants that were moved from one habitat to the other in order to find a suitable place for their maintenance) were scored based on growth in the habitat in which they performed best. These values have been show to constitute a meaningful proxy of the overall ability of species to grow and reproduce (Herben et al., 2012, 2014).
Data analysis
All analyses were done using R version 3.2.3 (R Core Team, 2015). To assess phylogenetic relationships of traits, we fitted phylogenetic least squares regressions using function ‘pgls’ from the package ‘caper’ for R (Orme, 2012). We always fitted two models: trait ∼ ploidy type and trait ∼ 1, calculated the adjusted R2 of the former and tested the significance of ploidy type as a predictor using F-statistics based on the comparison of the two models. We used regression coefficients for individual ploidy levels as an indicator of whether given ploidy contributes positively or negatively to the value of the given trait in the phylogenetic regression. Each pair of models was fitted for two different phylogenetic hypotheses: (1) a non-phylogenetic model with Pagel’s λ equal to zero (i.e. an ultrametric ‘star-like phylogeny’) and (2) a phylogenetic model with λ estimated from the data using maximum likelihood with the function ‘pgls’ as above. Pagel’s λ (Freckleton et al., 2002) is a multiplicative constant which expresses the role of internal structure of the tree in determination of values of the given trait. Its value used in phylogenetic regressions was estimated from the base model (trait ∼ 1).
Phylogenetic signals of response variable traits used in the regression were assessed using Pagel’s λ (Freckleton et al., 2002) by fitting the maximum likelihood model using the function ‘pgls’ from the package ‘caper’ for R (Orme, 2012). As reproduction scores are expressed on an ordinal scale, we also used generalized ordinal regression using the R package ‘ordinal’ (Christensen, 2011), but the results for all traits examined were very similar to those yielded by linear models. We therefore report only data from linear models.
Phylogenetic signal in the ploidy level was assessed using the D-statistics of Fritz and Purvis (2010), and we tested likelihood of the estimated value under two extreme hypotheses: (1) λ = 0 (no phylogenetic signal) and (2) λ = 1 (Brownian model of trait evolution over the phylogenetic tree, i.e. complete phylogenetic signal). The fitting and tests were done using the function ‘phylo.d’ from the package ‘caper’.
Phylogenetic relationships between ploidy and and traits of vegetative reproduction and reproduction scores were also modelled using the OU model assuming different trait optima (theta) for diploid and polyploid taxa. We assumed identical alpha (pull towards the optimum values) and sigma (random variation) for diploids and polyploids as simultaneous ploidy type-specific estimates of theta, and the variation parameters were unstable (assessed by eigenvalues of the Hessian matrix at the solution point) and there is no strong biological reason why they should differ between the ploidy types. We fitted the OU model using the function ‘OUwie’ from the R package ‘OUwie’ (Beaulieu and O’Meara, 2016; see also Beaulieu et al., 2012).
Finally, correlated evolution of ploidy and vegetative reproduction was also examined using the approach proposed by Pagel (1994; see, for example, Holden and Mace, 1997; Robertson et al., 2011) as implemented in the function ‘fitPagel’ (package ‘phytools’; Revell, 2012, 2014). While this approach may produce erroneous results under certain circumstances (‘unreplicated burst’ scenario of Maddison of FitzJohn, 2015), we believe that multiple reversals in all traits examined make it a suitable approach for addressing this question in our data, It models transition probabilities between character states along the phylogenetic tree either as independent of or dependent on each other and performs the test based on comparing likelihoods of these two models. We first fitted the two models (one assuming complete independence of evolution of each trait, and one assuming complete dependence) and compared them using the log likelihood test. No transition was forced to be zero in any of the models. Fitting of these models within the ‘fitPagel’ function was done using the function ‘fitDiscrete’ of the package ‘geiger’ (Harmon et al., 2014). As the Daphne phylogeny contains polytomies, these were converted to dichotomies by randomly assigning short (0·01 Mya) branch lengths to individual taxa; exploratory analyses showed that the outcome of the modelling did not differ among different random placements of these dichotomies. Further, to determine whether individual transition rates of one trait are dependent on the state of the other trait, we took a model with all transitions dependent (i.e. each of the eight transitions was allowed to vary) and compared its fit with the model with a model with one constraint placed on the transition being tested that made it independent of the value of the other trait. For example, when modelling transition to polyploidy in response to lateral spreading, we placed a constraint that the polyploidization rate be equal in plants with both long and short lateral spreading. We modelled evolution of polyploidy with three traits that showed the strongest relationship to ploidy in phylogenetic regressions, i.e. vegetative reproduction, bud bank size and lateral spreading distance. We divided each trait into two classes of approximately equal size to express it as a binary variable. We hence used the following cut-off points: bud bank size (19), vegetative reproduction (2) and lateral spreading (0·05 cm). Preliminary analysis showed that the results are essentially unaffected by the choice of these cut-off points.
RESULTS
All traits examined showed detectable phylogenetic conservatism. It was particularly high for some clonal traits (lateral spreading), lower for bud bank size and lowest for the number of clonal offspring (Table 2). Reproduction scores had generally weaker phylogenetic conservatism than clonal traits (Table 2). Ploidy type was also to some extent phylogenetically conservative: Fritz and Purvis’s D-statistics is 0·453 (note that values close to zero mean phylogenetic conservatism while values close to 1 mean phylogenetic independence).
Table 2.
Variation of reproduction scores and individual traits of clonal growth explained by the ploidy type
| Units | Lambda | CI lambda | Res.d.f. | Phylogenetic |
Star-like |
|||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Adjusted R2 | F | P-value | Difference | Adjusted R2 | F | P-value | Difference | |||||
| Clonal traits | ||||||||||||
| No. of clonal offspring | 1 | 0·594 | 0·371–0·757 | 453 | 0·002 | 1·75 | 0·187 | 0·241 | 0·005 | 3·13 | 0·078 | 0·317 |
| Lateral spreading distance | m | 0·856 | 0·734–0·919 | 453 | 0·029 | 14·71 | 0·000 | 0·027 | 0·012 | 6·68 | 0·010 | 0·019 |
| Bud bank size | 1 | 0·703 | 0·547–0·812 | 737 | 0·012 | 10·13 | 0·002 | 1·429 | 0·022 | 17·93 | 0·000 | 1·816 |
| Reproduction scores | ||||||||||||
| Seed reproduction | Ordinal (1–5) | 0·629 | 0·46–0·755 | 850 | 0·017 | 15·35 | 0·000 | –0·325 | 0·027 | 24·85 | 0·000 | –0·384 |
| Vegetative reproduction | Ordinal (1–5) | 0·687 | 0·529–0·797 | 893 | 0·022 | 21·42 | 0·000 | 0·435 | 0·050 | 47·97 | 0·000 | 0·623 |
Phylogenetic, phylogenetic regression with lambda estimated by maximum likelihood; Star-like, phylogenetic regression with lambda set to zero (i.e. least squares linear model).
Significant tests are given in bold.
Lambda, Pagel’s λ, CI lambda, 95 % confidence intervals of λ, Difference, regression coefficient of polyploids, i.e. estimated mean difference in the trait value of polyploids compared with diploids, expressed in units of the given trait.
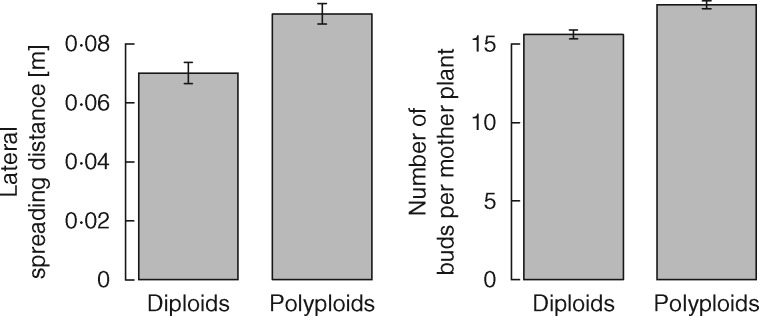
Out of clonal traits, bud bank size and lateral spreading distance showed significantly higher values for polyploids relative to diploids (Fig. 1; Table 2), while the number of clonal offspring per mother plant was not significant. Polyploids showed lateral spread on average by 2·7 cm more than diploids (in the phylogenetic model) and greater bud bank (1·8 buds; 1·5 buds in the phylogenetic model) than diploids (Table 2; Supplementary Data Fig. S1). Variation explained by ploidy type was generally lower in clonal traits than in reproduction scores (Table 2). These differences were highly significant (Table 2) even after correction for multiple testing.
Fig. 1.
Differences between diploids and polyploids) in lateral spreading distance and bud bank size (number of buds per mother plant). Error bars indicate the s.e.m. of each group.
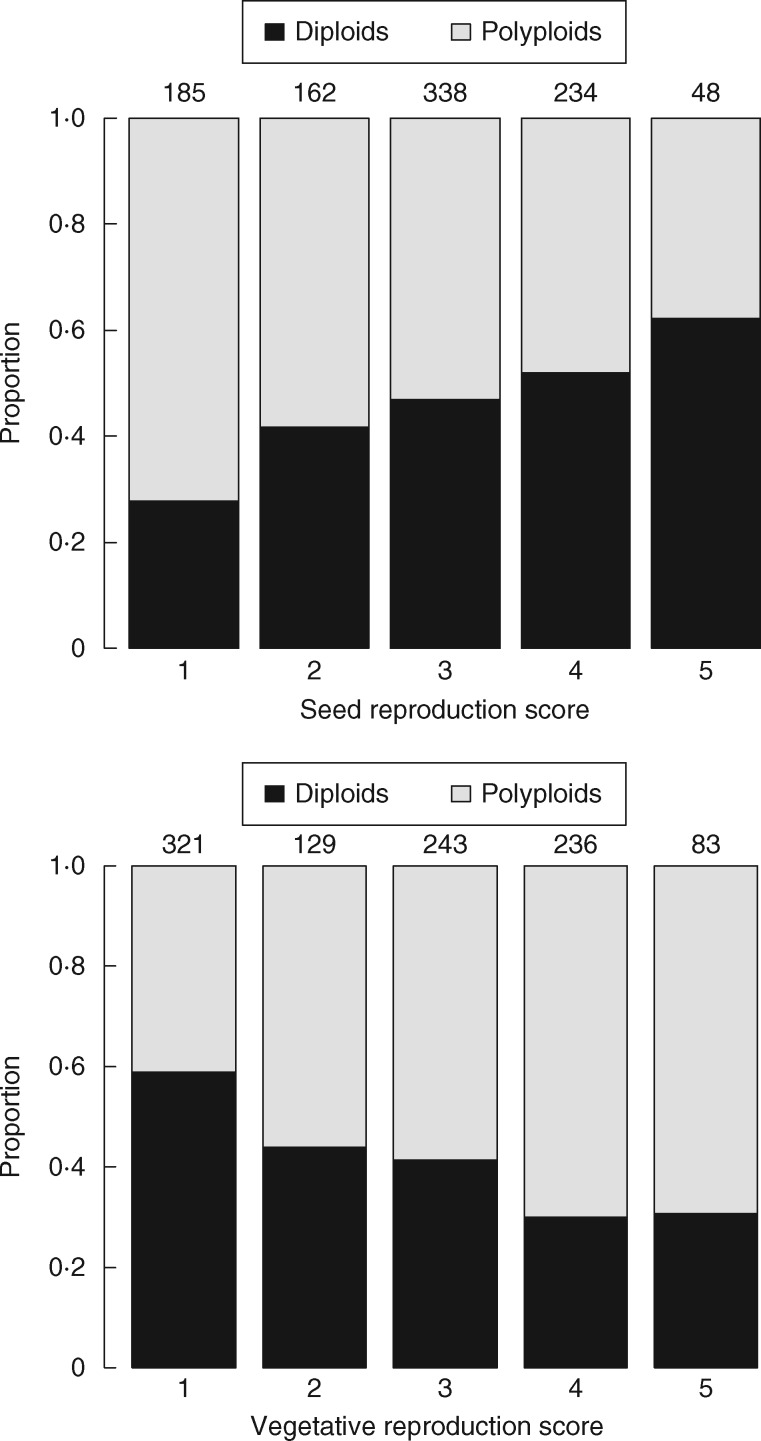
There were strong differences in the reproduction scores between individual ploidy types, with diploids showing the highest role of seed reproduction, and polyploids the highest vegetative reproduction (Fig. 2). On average, polyploids had a score of vegetative reproduction greater by 0·62 units, i.e. 15 % of the total range of the score (0·44 in phylogenetic analysis; Table 2). In contrast, their reproduction by seed was on average smaller by 0·38 units, i.e. 9·5 % of the total range. These differences were highly significant (Table 2). Differences between diploids and polyploids did not differ substantially between non-phylogenetic and phylogenetic analyses (Table 2).
Fig. 2.
Differences between diploids and polyploids in seed and vegetative reproduction score in the garden. Numbers indicate number of species in each group.
Models assuming the OU process showed significant differences between optimum values of diploids and polyploids in bud bank size (the optimum in polyploids is larger by two buds per individual; Table 3), lateral spreading distance (the optimum in polyploids is 4·7 cm larger) and vegetative reproduction score (the optimum in polyploids is 0·75 score units larger). In contrast, seed reproduction was significantly lower in polyploids (by 0·47 units). The number of clonal offspring had a higher optimum in polyploids, but the difference was not significant (95 % confidence intervals of the estimates overlap).
Table 3.
Estimates of optimum trait values for diploids and polyploids from the Ohrenstein–Uhlenbeck model
| Response | Units | Alpha | Sigma | Diploids |
Polyploids |
||
|---|---|---|---|---|---|---|---|
| Estimated optimum value | s.e. | Estimated optimum value | s.e. | ||||
| Clonal traits | |||||||
| No. of clonal offspring | 1 | 0·160 | 1·158 | 2·432 | 0·185 | 2·879 | 0·124 |
| Lateral spreading distance | m | 0·046 | 0·001 | 0·055 | 0·012 | 0·102 | 0·007 |
| Bud bank size | 1 | 0·135 | 9·419 | 15·030 | 0·412 | 17·079 | 0·327 |
| Reproduction scores | |||||||
| Seed reproduction | Ordinal (1–5) | 0·119 | 0·320 | 3·143 | 0·077 | 2·678 | 0·063 |
| Vegetative reproduction | Ordinal (1–5) | 0·101 | 0·376 | 2·053 | 0·094 | 2·804 | 0·075 |
Alpha, intensity of the selective regime; Sigma, error variation (in units2).
Significant differences between estimates of optimum values of diploids and polyploids are shown in bold.
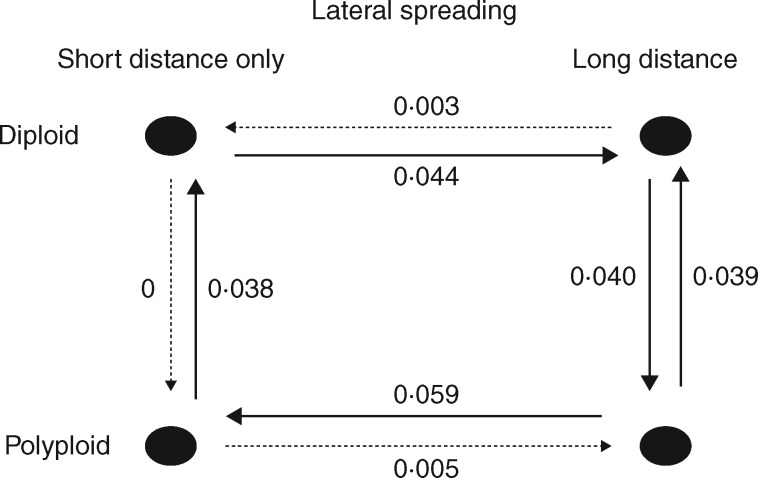
Models assuming dependence of polyploidization and appearance of vegetative reproduction always had significantly better fit than those assuming trait independence (Table 4, columns ‘global’). Tests of whether polyploidy appears more often in plants already having vegetative reproduction showed very strong conditioning of polyploidy by lateral spreading distance with no effect of other variables (Fig. 3; Table 4, columns P|V: 0|0 to 0|1 vs. 1|0 to 1|1). Tests of whether vegetative reproduction appears more often in polyploid plants invariably showed a negative effect, i.e. polyploid plants have a low chance of developing vegetative reproduction if it had not been present earlier (Fig. 3; Table 4, columns V|P: 0|0 to 1|0 vs. 0|1 to 1|1). Loss of vegetative reproduction may occur in polyploids, but it is very low in diploids (Fig. 3).
Table 4.
Tests of dependent evolution of polyploidy and vegetative reproduction
| Global | P-value | P|V | P-value | 0|0 to 0|1 | 1|0 to 1|1 | V|P | P-value | 0|0 to 1|0 | 0|1 to 1|1 | |
|---|---|---|---|---|---|---|---|---|---|---|
| LR | LR | LR | ||||||||
| Lateral spreading distance | 32·605 | 0·000 | 6·578 | 0·010 | 0·000 | 0·040 | 7·897 | 0·005 | 0·044 | 0·005 |
| Bud bank size | 23·850 | 0·000 | 14·092 | 0·000 | 0·040 | 0·002 | 2·581 | 0·108 | 0·096 | 0·047 |
| Vegetative reproduction score | 32·232 | 0·000 | 1·831 | 0·176 | 0·033 | 0·012 | 0·116 | 0·733 | 0·031 | 0·037 |
Overall test difference between a model with all transitions independent and a model with all transitions dependent; P|V reports tests of whether polyploidization depends on the presence of vegetative reproduction, V|P reports tests of whether the appearance of a given vegetative reproduction depends on polyploidy. 0|0 to 0|1, rate of polyploidization when vegetative reproduction is absent; 1|0 to 1|1, rate of polyploidization when vegetative reproduction is present; 0|0 to 1|0, rate of appearance of the particular type of vegetative reproduction in diploid plants; 0|1 to 1|1, rate of the appearance of vegetative reproduction in polyploid plants. LR, likelihood ratio (d.f. = 4 for the global test, d.f. = 1 for single transition tests). Significant tests are shown in bold. The last two columns provide a summary of the results based on a comparison of numerical values of coefficient pairs (0|0 to 0|1 vs. 1|0 to 1|1, and 0|0 to 1|0 vs. 0|1 to 1|1) and on associated significance values.
Fig. 3.
Rates of polyploidization and of appearance of long lateral spreading given different states of the other variable. Thick lines indicate relationships with rates > 0·03. Transition rates are estimated from the model with all parameters free (i.e. assuming no constraint on parameter values). Differences between the two polyploidization rates, and the two rates of appearance of lateral spreading are highly significant (Table 3).
DISCUSSION
Our data on nearly 1000 Angiosperm species from central Europe firmly support the hypothesis that polyploidy is associated with higher vegetative reproduction. This can be shown both by using values of two clonal traits (bud bank size and lateral spreading distance) and by actual vegetative reproduction assessed using long-term data from botanical garden collections. These differences can be identified independently of the underlying evolutionary model. Although the range of variation of all clonal traits within polyploid taxa and within diploid taxa is large (as indicated by the low proportion of explained variation in phylogenetic regressions), differences between means of both groups are large relative to ranges of the parameters tested and are very likely to be biologically meaningful.
However, our analyses also show that traits of clonal growth do not form a homogeneous group in their relationship to polyploidy. Specifically, the number of clonal offspring, as a proxy for clonal multiplication, did not differ between diploids and polyploids, implying that the rate of clonal reproduction is not the key trait associated with polyploidy. In contrast, the greatest differences between diploids and polyploids were found in the distance of lateral spreading. These differences can help to identify possible mechanism behind clonality and polyploidy: increasing lateral spreading distance places clonal offspring at greater distances, which may enhance their survival by reducing self-competition (Schmid and Harper, 1985) as in many species these distances are greater than their ramet height (J. Klimešová, unpubl. data). It also helps to encroach on space more efficiently, reducing mortality by small-scale disturbance events. Interestingly, in his pioneering analysis, Gustafsson (1948) implied that the important plant trait associated with polyploidy is the capability to spread by clonal growth, not the capability to resprout vegetatively (for which he probably did not have good data), a conclusion that is fully supported by our data.
A decrease in the explained variation in phylogenetic regression in comparison with star-like evolution implies that part of the variation in ploidy and clonal traits is due to shared evolutionary history (Freckleton et al., 2002), but even when this history is taken away, there is a substantial indication of correlated evolution of these two traits. This makes it possible to address explicitly the question of whether clonality is selected for and facilitated by polyploidy, or vice versa. Considerable phylogenetic signal in the traits analysed permits more detailed modelling of mutual (in)dependence of evolutionary rates of both processes, which showed convincingly that the observed patterns are primarily due to the evolution of polyploidy in species with vigorous vegetative growth, whereas the development of vegetative reproduction after a polyploidization event (either by direct effects of genome duplication or by selection associated with reduced sexual reproduction) was unlikely. Lateral spreading distance (i.e. the character with the greatest difference between diploids and polyploids) showed the strongest pattern of dependent evolution out of all traits tested; the chance of polyploid speciation in plants with high lateral spreading was two orders of magnitude higher than that in plants with no or short-distance lateral spreading. In contrast, once the species is polyploid, it can lose the ability for lateral spreading (presumably after overcoming the sterility barrier present immediately after polyploidization). Interestingly, the precedence of clonal reproduction over polyploidization was already suggested by Gustafsson (1948) and more recently discussed by Baldwin and Husband (2013). The latter authors found that genome duplication did not increase the expression of clonality in Chamerion angustifolium, indicating that polyploidy is a consequence, not the cause, of clonality. Actually, the precedence of clonality over genome duplication seems to be a general pattern in both plants and animals, as indicated by the results of Choleva et al. (2012). By reconstructing the entire evolutionary route from sexuality to clonality and polyploidy in the Cobitis fish, the authors provided direct evidence that clonality is directly triggered by inter-specific hybridization, and that polyploidy is a consequence, not the cause, of clonality in vertebrate animals.
Our results obtained at a large evolutionary scale seemingly contradict the data that come from detailed studies of individual polyploid complexes, in which the degree of clonality increases with increased ploidy level (e.g. Campanula patula: F. Rooks et al., unpubl. res.; Hieracium echioides: P. Trávníček et al., unpubl. res.). However, these case studies do not have the potential to demonstrate in which of these two variables change appeared earlier. Further, and more importantly, our analysis has coarser resolution over the phylogenetic tree and thus can address deeper differences in patterns of clonal growth. Some traits underlying clonal growth are considerably phylogenetically conserved (see, for example, Klimeš et al., 1997) and are likely not to differ within complexes of closely related species, where one would expect primarily quantitative differences such as modulation of the intensity of lateral growth. Therefore. it is conceivable that lineages with conserved absence of clonal traits are less prone to polyploid speciation (e.g. Apiaceae), whereas lineages where traits of vegetative reproduction routinely appear may use this potential further and even increase it after a polyploidization event (e.g. many aquatic groups).
It should be noted that our analyses are based on a fairly uneven sampling of the plant phylogeny, and we possess no information on whether and how the relationships reported would change for larger sets of species. However, our taxon sampling is limited to the only flora for which comparative information on clonal traits and vegetative reproduction exists, and in this respect knowledge of the flora of Central Europe is by far better than that of any other region of the world. However, while our sample list includes only temperate taxa, Central Europe is ecologically fairly strongly differentiated, ranging from lowlands to mountains, and consequently the taxon sampling represents a reasonable range of ecological strategies of plants in several major clades. In addition, the fairly narrow geographic focus rules out possible alternative explanations that the results are a by-product of separate correlations between traits and latitude.
Our study of a representative sample of central European flora contributed to better understanding of the evolutionary significance of whole-genome duplication, which is arguably the single most important genetic mechanism in plant speciation and diversification. The data presented here provide a strong argument for the classical hypothesis that polyploidy is associated with increased vegetative reproduction (Gustafsson, 1948). In addition, we tackled the question of whether polyploidy is the cause or the consequence of clonality (see Baldwin and Husband, 2013), supporting the latter possibility. In order to determine the generality of our conclusions and to explore fully the link between polyploidy and clonal reproduction, future research should involve representatives of other floras, assess potential differences between different types of polyploids (i.e. auto- vs. allopolyploids) and compare naturally occurring polyploids with their synthetic counterparts.
SUPPLEMENTARY DATA
Supplementary data are available online at https://academic.oup.com/aob and consist of the following. Table S1: data used in the analyses. Table S2: correlation matrix of predictor variables. Figure S1: phylogenetic trees with vegetative reproduction score and lateral spreading distance mapped together with ploidy status
Supplementary Material
ACKNOWLEDGEMENTS
Analysis of reproductive patterns in the garden would be impossible without the long-term dedicated work of Zuzana Nováková in the Botanical Garden of Charles University. We thank Brian Husband who first pointed out the possibility of testing precedence of clonal reproduction by phylogenetic techniques. We further thank Petr Šmarda for the newest version of the phylogenetic tree of the Czech flora, Liam Revell for advice on the phytools package, and two anonymous referees for constructive comments on an earlier version of the paper. The research was partly supported by the Grant Agency of the Czech Republic (project GA 16-19245S). Jan Suda passed away on 9th March 2017.
LITERATURE CITED
- Asker SE, Jerling L.. 1992. Apomixis in plants. Boca Raton, FL: CRC Press. [Google Scholar]
- Baldwin SJ, Husband BC.. 2013. The association between polyploidy and clonal reproduction in diploid and tetraploid Chamerion angustifolium. Molecular Ecology 22: 1806–1819. [DOI] [PubMed] [Google Scholar]
- Bennett MD, Leitch IJ.. 2012. Pteridophyte DNA C-values database (Release 5.0, December 2012). http://data.kew.org/cvalues.
- Beaulieu JM, O’Meara B.. 2016. OUwie: analysis of evolutionary rates in an OU framework. R package version 1·50. https://CRAN.R-project.org/package=OUwie.
- Beaulieu JM, Jhwueng DC, Boettiger C, O’Meara BC.. 2012. Modeling stabilizing selection: expanding the Ornstein–Uhlenbeck model of adaptive evolution. Evolution 66: 2369–2383. [DOI] [PubMed] [Google Scholar]
- Boedeltje G, Ozinga WA, Prinzing A.. 2008. The trade-off between vegetative and generative reproduction among angiosperms influences regional hydrochorous propagule pressure. Global Ecology and Biogeography 17: 50–58. [Google Scholar]
- Choleva L, Janko K, De Gelas K, et al. 2012. Synthesis of clonality and polyploidy in vertebrate animals by hybridization between two sexual species. Evolution 66: 2191–2203. [DOI] [PubMed] [Google Scholar]
- Christensen RHB. 2011. Ordinal regression models for ordinal data. R package version 2010.10-22 http://www.cran.r-project.org/package=ordinal/.
- Comai L. 2005. The advantages and disadvantages of being polyploid. Nature Reviews Genetics 6: 836–846. [DOI] [PubMed] [Google Scholar]
- Dobeš C, Vitek E.. 2000. Documented chromosome number checklist of Austrian vascular plants. Wien, Österreich: Verlag des Naturhistorischen Museums Wien. [Google Scholar]
- Doležel J, Greilhuber J, Suda J.. 2007. Flow cytometry with plants: an overview In: Doležel JJ, Greilhuber J, Suda J, eds. Flow cytometry with plant cells. Analysis of genes, chromosomes and genomes. Weinheim, Germany: Wiley-VCH, 41–65. [Google Scholar]
- Duchoslav M., Staňková H.. 2015. The population genetic structure and clonal diversity of Allium oleraceum (Amaryllidaceae), a polyploid geophyte with common asexual but variable sexual reproduction. Folia Geobotanica 50: 123–136. [Google Scholar]
- Durka W, Michalski SG.. 2012. Daphne: a dated phylogeny of a large European flora for phylogenetically informed ecological analyses. Ecology 93: 2297–2297. [Google Scholar]
- Eckert CG, Lui K, Bronson K, Corradini P, Bruneau A.. 2003. Population genetic consequences of extreme variation in sexual and clonal reproduction in an aquatic plant. Molecular Ecology 12: 331–344. [DOI] [PubMed] [Google Scholar]
- Felber F. 1991. Establishment of a tetraploid cytotype in a diploid population: effect of relative fitness of the cytotypes. Journal of Evolutionary Biology 4: 195–207. [Google Scholar]
- Fowler NL, Levin DA.. 1984. Ecological constraints on the establishment of a novel polyploid in competition with its diploid progenitor. American Naturalist 124: 701–711. [Google Scholar]
- Freckleton RP, Harvey PH, Pagel M.. 2002. Phylogenetic analysis and comparative data: a test and review of evidence. American Naturalist 160: 712–26. [DOI] [PubMed] [Google Scholar]
- Fritz SA, Purvis A.. 2010. Selectivity in mammalian extinction risk and threat types: a new measure of phylogenetic signal strength in binary traits. Conservation Biology 24: 1042–1051. [DOI] [PubMed] [Google Scholar]
- Glick L, Mayrose I.. 2014. ChromEvol: assessing the pattern of chromosome number evolution and the inference of polyploidy along a phylogeny. Molecular Biology and Evolution 31: 1914–1922. [DOI] [PubMed] [Google Scholar]
- Goldblatt P. 1980. Polyploidy in angiosperms: monocotyledons In: Lewis WH, ed. Polyploidy: biological relevance. New York: Plenum Press, 219–239. [Google Scholar]
- Goldblatt P, Johnson DE.. 1979. Index to plant chromosome numbers. St Louis, MO: Missouri Botanical Garden. [Google Scholar]
- Góralski G, Lubczyńska P, Joachimiak AJ.. 2009. onwards. Chromosome number database http://www.chromosomes.binoz.uj.edu.pl/chromosomes/.
- Grant V. 1981. Plant speciation, 2nd edn New York: Columbia University Press. [Google Scholar]
- Gustafsson A. 1948. Polyploidy, life-form and vegetative reproduction. Hereditas 34: 1–22. [Google Scholar]
- Harmon L, Weir J, Brock C, et al. , 2014. Package ‘geiger’ https://cran.r-project.org/web/packages/geiger.
- Herben T, Nováková Z, Klimešová J, Hrouda L.. 2012. Species traits and plant performance: functional trade-offs in a large set of species in a botanical garden. Journal of Ecology 100: 1522–1533. [Google Scholar]
- Herben T, Nováková Z, Klimešová J.. 2014. Clonal growth and plant species abundance. Annals of Botany 114: 377–388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holden C, Mace R.. 1997. Phylogenetic analysis of the evolution of lactose digestion in adults. Human Biology 69: 605–628. [PubMed] [Google Scholar]
- Husband BC. 2004. The role of triploid hybrids in the evolutionary dynamics of mixed-ploidy populations. Biological Journal of the Linnean Society 82: 537–546. [Google Scholar]
- Husband BC, Baldwin SJ, Suda J.. 2013. The incidence of polyploidy in natural plant populations: major patterns and evolutionary processes In: Leitch IJ, Greilhuber J, Doležel J, Wendel JF, eds. Plant genome diversity Volume 2. Physical structure, behaviour and evolution of plant genomes. Wien, Austria: Springer Verlag, 255–276. [Google Scholar]
- Jiao Y, Wickett NJ, Ayyampalayam S, et al. 2011. Ancestral polyploidy in seed plants and angiosperms. Nature 473: 97–100. [DOI] [PubMed] [Google Scholar]
- Keeler KH. 2004. Impact of intraspecific polyploidy in Andropogon gerardii (Poaceae) populations. American Midland Naturalist 152: 63–74. [Google Scholar]
- Klimeš L, Klimešová J, Hendriks R, van Groenendael J.. 1997. Clonal plant architecture: a comparative analysis of form and function In: de Kroon H, van Groenendael J, eds. The ecology and evolution of clonal plants. Leiden, The Netherlands: Backhuys Publishers, 1–29. [Google Scholar]
- Klimešová J, de Bello F.. 2009. CLO-PLA: the database of clonal and bud bank traits of Central European flora. Journal of Vegetation Science 20: 511–516. [Google Scholar]
- Klimešová J, Tackenberg O, Herben T.. 2016. Herbs are different: clonal and bud bank traits can matter more than leaf–height–seed traits. New Phytologist 210: 13–17. [DOI] [PubMed] [Google Scholar]
- Levin D. 1975. Minority cytotype exclusion in local plant populations. Taxon 24: 35–43. [Google Scholar]
- Lososová Z, Šmarda P, Chytrý M, et al. 2015. Phylogenetic structure of plant species pools reflects habitat age on the geological time scale. Journal of Vegetation Science 26: 1080–1089. [Google Scholar]
- Maddison WP, FitzJohn RG.. 2015. The unsolved challenge to phylogenetic correlation tests for categorical characters. Systematic Biology 64: 127–136. [DOI] [PubMed] [Google Scholar]
- Marhold P, Mártonfi P, Mereďa P, et al. 2012. Karyological database of the ferns and flowering plants of Slovakia.http://www.chromosomes.sav.sk.
- Meyers LA, Levin DA.. 2006. On the abundance of polyploids in flowering plants. Evolution 60: 1198–1206. [PubMed] [Google Scholar]
- Obeso JR. 2002. The costs of reproduction in plants. New Phytologist 155: 321–348. [DOI] [PubMed] [Google Scholar]
- Orme D. 2012. The Caper package: comparative analysis of phylogenetics and evolution in R http://cran.r-project.org/web/packages/caper/.
- Otto SP, Whitton J.. 2000. Polyploid incidence and evolution. Annual Review of Genetics 34: 401–437. [DOI] [PubMed] [Google Scholar]
- Pagel M. 1994. Detecting correlated evolution on phylogenies: a general method for the comparative analysis of discrete characters. Proceedings of the Royal Society B: Biological Sciences 255: 37–45. [Google Scholar]
- R Core Team. 2015. R: a language and environment for statistical computing R Foundation for Statistical Computing, Vienna, Austria. https://www.R-project.org/.
- Ramsey J, Schemske DW.. 1998. Pathways, mechanisms and rates of polyploidy formation in flowering plants. Annual Review of Ecology and Systematics 29: 467–501. [Google Scholar]
- Revell LJ. 2012. phytools: an R package for phylogenetic comparative biology (and other things). Methods in Ecology and Evolution 3: 217–223. [Google Scholar]
- Revell LJ. 2014. Package ‘phytools’.https://cran.r-project.org/web/packages/phytools.
- Reznick D, Nunney L, Tessier A.. 2000. Big houses, big cars, superfleas and the costs of reproduction. Trends in Ecology and Evolution 15: 421–425. [DOI] [PubMed] [Google Scholar]
- Rice A, Glick L, Abadi S, et al. 2015. The Chromosome Counts Database (CCDB) – a community resource of plant chromosome numbers. New Phytologist 206: 19–26. [DOI] [PubMed] [Google Scholar]
- Robertson K, Goldberg EE, Igic B.. 2011. Comparative evidence for the correlated evolution of polyploidy and self-compatibility in Solanaceae. Evolution 65: 139–155. [DOI] [PubMed] [Google Scholar]
- Schmid B, Harper JL.. 1985. Clonal growth in grassland perennials I. Density and pattern dependent competition between plants with different growth forms. Journal of Ecology 73: 793–808. [Google Scholar]
- Soltis DE, Buggs RJA, Doyle JJ, Soltis PS.. 2010. What we still don’t know about polyploidy. Taxon 59: 1387–1403. [Google Scholar]
- Soltis DE, Segovia-Salcedo MC, Jordon-Thaden I, et al. 2014. Are polyploids really evolutionary dead-ends (again)? A critical reappraisal of Mayrose et al. (2011). New Phytologist 202: 1105–1117. [DOI] [PubMed] [Google Scholar]
- Stebbins GL. 1957. Self fertilization and population variability in the higher plants. American Naturalist 91: 337–354. [Google Scholar]
- Weiss-Schneeweiss H, Emadzade K, Jang T-S, Schneeweiss GM.. 2013. Evolutionary consequences, constraints, and potential of polyploidy in plants. Cytogenetic and Genome Research 140: 137–150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Wettstein F. 1927. Die Erscheinung der Heteroploidie, besonders im Pflanzenreich. Ergebnise der Biologie, Bd. 11. Berlin: Springer Verlag. [Google Scholar]
- Whitton J, Sears CJ, Baack EJ, Otto SP.. 2008. The dynamic nature of apomixis in the angiosperms. International Journal of Plant Sciences 169: 169–182. [Google Scholar]
- Wood TE, Takebayashi N, Barker MS, Mayrose I, Greenspoon PB, Rieseberg LH.. 2009. The frequency of polyploidy speciation in plants. Proceedings of the National Academy of Sciences, USA 106: 13875–13879. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.