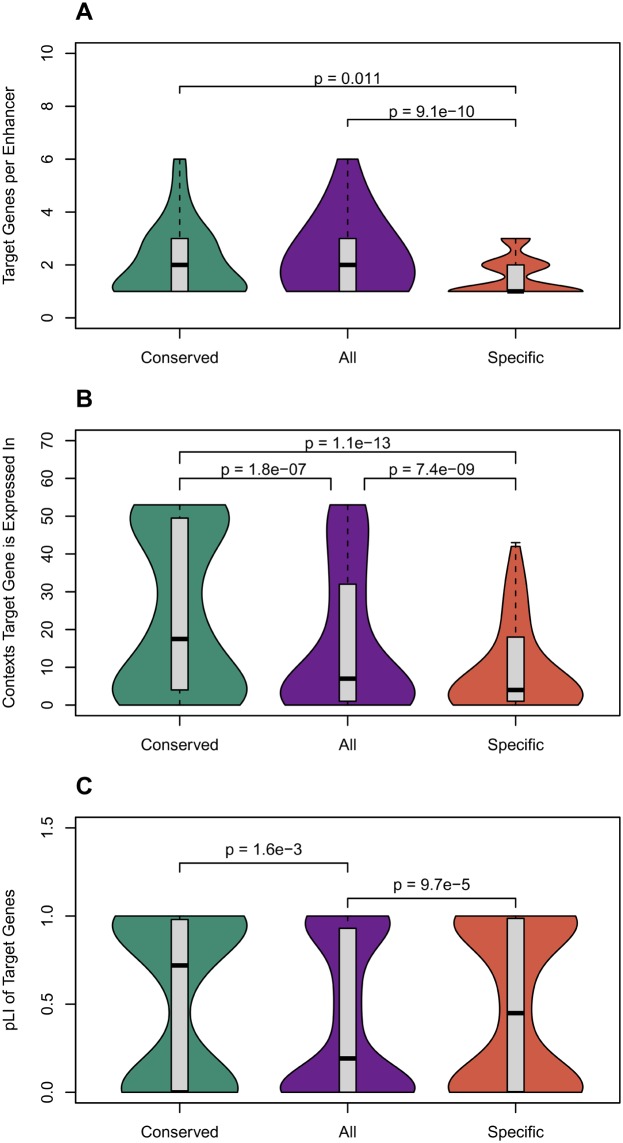
Fig. 5.
—Conserved-activity enhancers have more target genes than human-specific-activity enhancers; their targets are expressed in more cellular contexts; and their targets are under stronger evolutionary constraint. (A) Conserved-activity enhancers have significantly more target genes than human-specific-activity enhancers, but a similar number as all human liver enhancers. (B) The gene targets of conserved-activity enhancers were active in significantly more contexts than either human-specific-activity or all liver enhancers. Target gene expression was determined from GTEx (The GTEx Consortium 2015). (C) The target genes for conserved-activity enhancers have significantly higher probabilities of being loss-of-function intolerant (pLI) according to ExAC than target genes for all human liver enhancers. Thus, the target genes of conserved-activity enhancers are under more evolutionary constraint. The targets of human-specific-activity enhancers were also less tolerant of loss of function than liver enhancers overall. Enhancers were mapped to target genes based on coexpression patterns by the FANTOM Consortium. Results were similar when identifying targets based on eQTL (supplementary fig. 6, Supplementary Material online ). The Mann–Whitney U test was used for all comparisons. Distributions are summarized as described for figure 2.