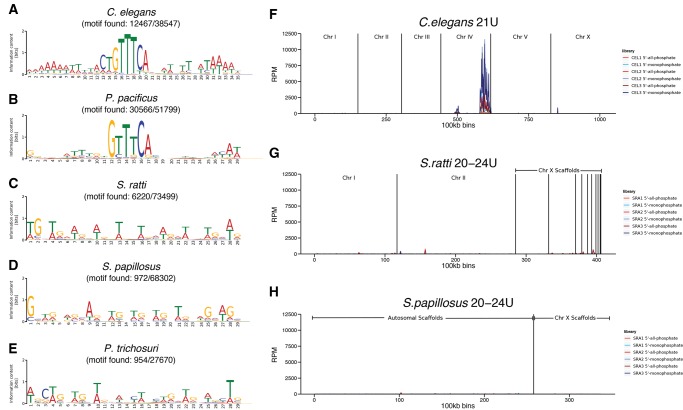
Fig. 5.
—Absence of a Ruby-type motif and no genomic clustering of 20–24 U RNAs in Strongyloididae. (A–E) Sequence logos of de novo predicted motifs based on 5,000 randomly selected 60 bp regions immediately upstream of aligned 21 U (A, B) or 20–24 U (C–E) other-mapped sequences. The numbers beneath the species indicate the number of places in the genome to which 21 U or 20–24 U reads align that do have the motif in the upstream region/total number of places to which 21 U or 20–24 U reads align. (F–H) Plots representing the genome coverage by 21 U or 20–24 U other-mapped reads from adult worm replicates along the chromosomes/scaffolds (100kb bins) for C. elegans (F), S. ratti (G), and S. papillosus (H). The different biological replicates and treatments are color coded. The two strongest peaks in C. elegans correspond to the known major and minor piRNA gene clusters. RPM reads per million.