Abstract
Genome remodeling and exchange of sequences are widespread in the prokaryotic world and mosaic genomes challenge the classification of prokaryotes, which cannot be properly achieved in terms of a single gene or group of genes. Here, we studied individually the gene collection of the archaic microorganism Lokiarchaeum sp., suggested as an archaeal host close to the emergence of the eukaryotes. The network or rhizome of all Lokiarchaeum sp. genes revealed that the genomic repertoire is mainly composed of genes from archaeal (∼36%) and bacterial origin (∼28%), distantly followed by components of eukaryotic origin (∼2%). Thirty-three percent of genes were unique to this species (ORFans). The mosaicity of archaea was also supported by studying Methanomassiliicoccus luminyensis, an archaea from the gut, in which 67% of the genomic repertoire arised from archaea and 22% from bacteria. Our results illustrate the intricate evolutionary relationships of the archaeal genome repertoire and highlight the rhizome-like processes of evolution in archaea, their mosaicity, and chimeric origin composed of different domains of life, questioning the reality of a tree of life.
Keywords: tree of life, Lokiarchaeota, gene exchange, mosaic genomes
Darwin used metaphor from the bible such as the tree of life, in which each single branch emerges from a common ancestor (nowadays referred as Last Universal Common Ancestor, LUCA). LUCA, as our “Adam” like organism, is not realistic now (Raoult and Koonin 2012; Raoult 2013). The initial and extensive use of ribosomal RNA as the gold-standard taxonomic molecular marker leads to consideration of three distinct domains of life: Eukaryota, Archaea, and Bacteria (Woese and Fox 1977; Woese 1987,Woese etal. 1990). However, classification based on a single gene or group of genes raises the problem of organism homogenicity. Consequently, the tree of life and the domain definition are based on a small fraction of the genome, involving only 1% of available data (Doolittle 1999, 2009a; Wolf etal. 2002; Dagan and Martin 2006; Doolittle and Bapteste 2007). The tree topology based on the ribosome suggests that Eukaryota and Archaea share a common ancestral set of ribosomal genes. Accordingly, the genomic description of putative archaic microorganisms such as Lokiarchaeota (Spang etal. 2015), in which numerous genes are found both in eukaryotes and archaea, leads to classification of this microorganism as a possible ancestor of Eukaryota and Archaea which is a biological Graal. Alternatively, it has also been proposed that modern microorganisms emerged from a profound “rhizome,” “bushes,” or “a network,” in order to explain the multiple origins of eukaryote and archaeal genes (Gould 1987; Gogarten etal. 2002; Gogarten and Townsend 2005; Raoult 2010a; Dunning Hotopp 2011; Koonin etal. 2011; Merhej etal. 2011; Georgiades and Raoult 2012). Here, we compared sequence exchanges between the archaic Lokiarchaeota and eukaryotes, as well as archaea and bacteria living in the gut in the same common ecosystem. Our objectives are to reassess the data of Spang etal. at the light of the study of the whole ORFeomes and to evaluate the mosaicity of archaeal genomes.
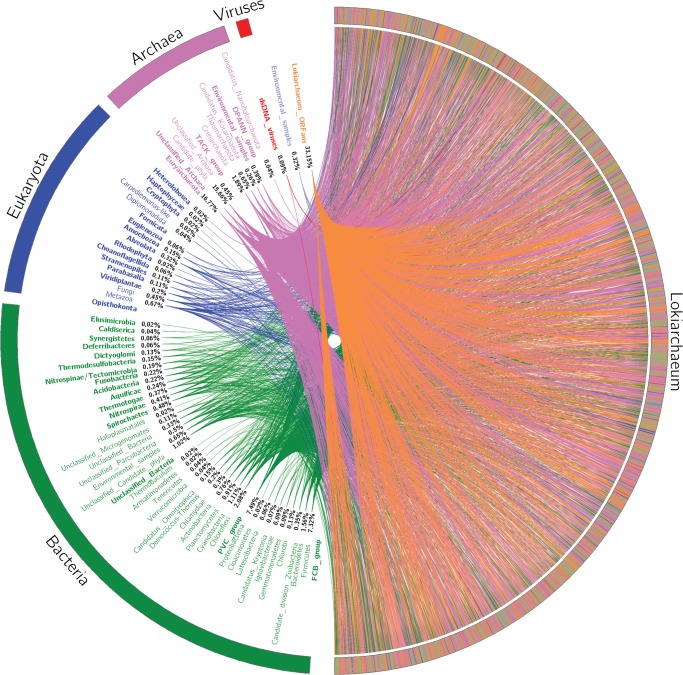
The discovery of a novel archaeal phylum candidate, Lokiarchaeota, was evidenced by Spang etal. (2015). The authors described this new archaeal lineage as forming a monophyletic group with eukaryotes and hypothesized a Lokiarchaeota–Eukarya affiliation and a putative missing link in the origin of organisms. The origin of all Lokiarchaeota genes was determined according to their taxonomic affiliation. Among the 5,384 Lokiarchaeota hits, 1,785 genes were unique to this species (fig. 1). The remaining genes were assigned to the different domains of life as follows: 1,944 genes (36.1%) in archaea, 1,509 genes (28%) in bacteria, and 121 genes (2.2%) in eukaryota (supplementary table 1, Supplementary Material online). Thus, the genome of Lokiarchaeota possesses a genomic repertoire mainly composed of genes from bacterial and archaeal origin, distantly followed by components of eukaryotic origin. The whole genome analysis confirmed our first results based on molecular markers and definitely placed Lokiarchaeum as a model of a mosaic genome composed of mainly bacterial and archaeal components, followed by eukaryote components.
Fig. 1.
—The rhizome of Lokiarchaeota. Viruses are depicted in red, archaea in pink, eukaryota in blue, and bacteria in green. ORFans are depicted in orange. All protein sequences were used as queries in a BLASTp search (Altschul etal. 1990) against the nonredundant (nr) protein database from NCBI. Blast results were filtered to keep the best hits and taxonomic affiliation was retrieved from NCBI. Best hit was selected and integrated in a circular gene data image (Krzywinski etal. 2009). The whole coding sequences of Lokiarchaeum was downloaded from NCBI, Lokiarchaeum sp. GC14_75 (PRJNA259156, JYIM01000001: JYIM01000504).
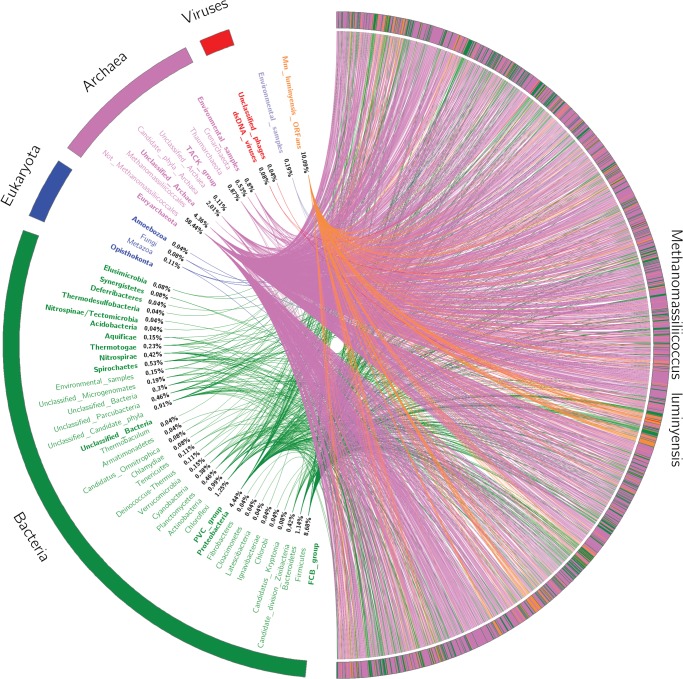
To illustrate the mosaicity of archaea, we also studied an archaea from the gut, M. luminyensis (Dridi etal. 2012). Sixty-seven percent of the genomic repertoire of M. luminyensis arised from archaea and 22% from bacteria (fig. 2 and supplementary table 1, Supplementary Material online). These results reveal the mosaic genome content of M. luminyensis. Altogether, these results are in accordance with recent findings that suggest gene exchange between bacteria and archaea living in the same ecological niche (Deschamps etal. 2014; Nelson-Sathi etal. 2015).
Fig. 2.
—The rhizome of Methanomassiliicoccus luminyensis. Viruses are depicted in red, archaea in pink, eukaryota in blue, and bacteria in green. ORFans are depicted in orange. The Methanomassiliicoccus luminyensis genome (NZ_CAJE00000000.1) was retrieved from NCBI (Dridi etal. 2012).
Our analysis of the gene content showed that archaeal genomes are rather of mosaic structure (Koonin and Yutin 2014). A significant part of the genome does not contain proteins of archaeal origin, but is constituted of bacterial proteins. Extensive DNA exchange and recombination lead, or at least contributes, to chimeric genomes. The composition of the genomes reflects its environment; in other terms, “you are what you eat,” “tell me where you live, I will tell you what you are.” Because of the mosaics of genomes, the construction of a Tree of Life (TOL) is rendered very difficult. For instance, the standard molecular marker 16 S rRNA gene could not be used, because several distinct copies of different 16S variants are found in a single species (Viezens and Arvand 2008). All attempts to find the origin of all life forms according to a simple LUCA vision, from which all living organisms have evolved, is probably misleading (Pace 2006). In addition, simulations of genes and organismal lineages suggest that there was no single common ancestor that contained all the genes ancestral to those shared among the three domains of life (Zhaxybayeva and Gogarten 2004). The last universal common ancestor of genes spread out over multiple organismal lineages, and existing at different times (Zhaxybayeva and Gogarten 2004). Some alternative strategies to build a TOL were based on molecular features that are not gene or proteins sequences. For instance, insertion and deletion (indel)-based investigation are applicable for rooting the TOL (Lake etal. 2007, 2009). The TOL controversy finds a striking illustration in the “bridge that gap between prokaryotes and eukaryotes” constructed from a concatenation of sequence alignments of highly conserved proteins, primarily those involved in translation, as illustrated in Lokiarchaeota (Spang etal. 2015). However, eukaryotes are rather chimeric organisms, with an archaeal subset that is strongly enriched in information processing functions (replication, transcription, and translation), a bacterial subset that consists largely of operational functions (metabolic enzymes and membrane proteins), endogenous retroviruses, and eukaryote-specific genes (Belshaw etal. 2004; Esser 2004; Rivera and Lake 2004; Yutin etal. 2008). Moreover, the purpose of concatenation is to reinforce weak phylogenetic signals that cannot be robustly recovered from any single gene with a very low bootstrap, but it is not appropriate, because the genes used have different phylogenetic stories that preclude concatenation (Rokas etal. 2003). Taking into account the universally distributed genes among all genomes and excluding lateral transfer, we may consider that on an average only 0.1% of a large eukaryotic proteome and 1% of a prokaryotic proteome fit the concept of a tree of life; that is, a common origin of all information encoded in organisms (Dagan and Martin 2006; Dagan etal. 2008). The representation of the TOL from Spang etal. (2015) can be summarily dismissed as a “tree of 1%” (of the genes in any given genome), which cannot represent genuine evolution.
Our result revealed the mosaic nature of the Lokiarchaeota genome mainly composed of genes from archaeal and bacterial origin. The high proportion of bacterial genes in Lokiarchaeota is in agreement with the interdomain gene transfer between archae and bacteria in which transfers from bacteria to archaea are 5-fold more frequent than vice versa (Nelson-Sathi etal. 2015).
In our study, BLAST approach used for taxonomic assignment could have phenetic shortcut (Koski and Golding 2001; Dick etal. 2017) as compared with the classical phylogenetic analyses. We confirmed the accuracy of the BLAST method by analyzing a subset of randomly selected Lokiarchaeota genes. Although, more precise and detailed, the phylogenetic approaches gave comparable results as compared with the BLAST approaches. Inferring the phylogenetic relationships between organisms definitely cannot be achieved by using a unique tree, due to the different origins of genes and subsequent dissimilar tree topologies (noncongruence). Moreover, although lateral gene transfer accounts for similarities between the domains of life, vertical inheritance from the common ancestor followed by loss of genes in one domain is still possible.
As pointed by our analysis of Lokiarchaeota or M. luminyensis, evolution should resemble a clump of roots representing the numerous origins of the genes repertoires for each species (Georgiades and Raoult 2012). Thus, our study suggests that a rhizome, forest, or bushes of genes representation is much more adequate to stand for the multiplicity and de novo creation of a genome. In addition, a rhizome applies to the entire cellular life history, contrary to the TOL. The TOL is based on scientific hypothesis not demonstrated and strongly reflecting the social beliefs during the Darwin’s period, as a direct resurgence of the creation theory (Raoult 2010b). The hierarchical order originating from a single ancestor is therefore erroneous and the universal scope of such tree-based evolutionary explanations has now been strongly questioned (Doolittle 1999, 2009a, 2009b; Bapteste etal. 2009; Dagan and Martin 2009). Microbial genome could be viewed as a mosaic of genes composed of eukaryotic, bacterial, archaeal, and viral genes that comprise an ecosystem enabling to drive a dynamic network of gene exchanges.
In conclusion, the novel phylum of Lokiarchaeota did not recover the “lost ark” of the TOL, but illustrates once again the rhizome-like processes of evolution (Raoult 2010a; Merhej etal. 2011) and rules out the existence of LUCA and the vertical evolution of the Neo-Darwinists. Eukaryotes remain complex mosaic organisms that possess genes of apparent archaeal origin, genes of probable bacterial origin, genes of viral origin, genes that seem to be eukaryote-specific and newly created genes, and there are no currently identified remains of the putative Archaea-Eukaryotes’ common ancestor.
Supplementary Material
Supplementary data are available at Genome Biology and Evolution online.
Supplementary Material
Literature Cited
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ.. 1990. Basic local alignment search tool. J Mol Biol. 215(3):403–410. [DOI] [PubMed] [Google Scholar]
- Bapteste E, et al. 2009. Prokaryotic evolution and the tree of life are two different things. Biol Direct. 4:34.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belshaw R, et al. 2004. Long-term reinfection of the human genome by endogenous retroviruses. Proc Natl Acad Sci U S A. 101(14):4894–4899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dagan T, Artzy-Randrup Y, Martin W.. 2008. Modular networks and cumulative impact of lateral transfer in prokaryote genome evolution. Proc Natl Acad Sci U S A. 105(29):10039–10044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dagan T, Martin W.. 2006. The tree of one percent. Genome Biol. 7(10):118.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dagan T, Martin W.. 2009. Getting a better picture of microbial evolution en route to a network of genomes. Philos Trans R Soc Lond B Biol Sci. 364(1527):2187–2196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deschamps P, Zivanovic Y, Moreira D, Rodriguez-Valera F, López-García P.. 2014. Pangenome evidence for extensive interdomain horizontal transfer affecting lineage core and shell genes in uncultured planktonic thaumarchaeota and euryarchaeota. Genome Biol Evol. 6(7):1549–1563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dick AA, Harlow TJ, Peter Gogarten J.. 2017. Short branches lead to systematic artifacts when BLAST searches are used as surrogate for phylogenetic reconstruction. Mol Phylogenet Evol. 107:338–344. [DOI] [PubMed] [Google Scholar]
- Doolittle WF. 1999. Phylogenetic classification and the universal tree. Science 284(5423):2124–2129. [DOI] [PubMed] [Google Scholar]
- Doolittle WF. 2009a. Eradicating typological thinking in prokaryotic systematics and evolution. Cold Spring Harb Symp Quant Biol. 74:197–204. [DOI] [PubMed] [Google Scholar]
- Doolittle WF. 2009b. The practice of classification and the theory of evolution, and what the demise of Charles Darwin’s tree of life hypothesis means for both of them. Philos Trans R Soc Lond B Biol Sci. 364(1527):2221–2228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doolittle WF, Bapteste E.. 2007. Pattern pluralism and the Tree of Life hypothesis. Proc Natl Acad Sci U S A. 104(7):2043–2049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dridi B, Fardeau ML, Ollivier B, Raoult D, Drancourt M.. 2012. Methanomassiliicoccus luminyensis gen. nov., sp. nov., a methanogenic archaeon isolated from human faeces. Int J Syst Evol Microbiol. 62(Pt 8):1902–1907. [DOI] [PubMed] [Google Scholar]
- Dunning Hotopp JC. 2011. Horizontal gene transfer between bacteria and animals. Trends Genet. 27(4):157–163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esser C. 2004. A genome phylogeny for mitochondria among alpha-proteobacteria and a predominantly eubacterial ancestry of yeast nuclear genes. Mol Biol Evol. 21(9):1643–1660. [DOI] [PubMed] [Google Scholar]
- Georgiades K, Raoult D.. 2012. How microbiology helps define the rhizome of life. Front Cell Infect Microbiol. 2:60.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gogarten JP, Doolittle WF, Lawrence JG.. 2002. Prokaryotic evolution in light of gene transfer. Mol Biol Evol. 19(12):2226–2238. [DOI] [PubMed] [Google Scholar]
- Gogarten JP, Townsend JP.. 2005. Horizontal gene transfer, genome innovation and evolution. Nat Rev Microbiol. 3(9):679–687. [DOI] [PubMed] [Google Scholar]
- Gould SJ. 1987. The empire of the apes. Nat Hist. 96:20. [Google Scholar]
- Koonin EV, Puigbò P, Wolf YI.. 2011. Comparison of phylogenetic trees and search for a central trend in the “forest of life”. J Comput Biol. 18(7):917–924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koonin EV, Yutin N.. 2014. The dispersed archaeal eukaryome and the complex archaeal ancestor of eukaryotes. Cold Spring Harb Perspect Biol. 6(4):a016188.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koski LB, Golding GB.. 2001. The closest BLAST hit is often not the nearest neighbor. J Mol Evol. 52(6):540–542. [DOI] [PubMed] [Google Scholar]
- Krzywinski M, et al. 2009. Circos: an information aesthetic for comparative genomics. Genome Res. 19(9):1639–1645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lake JA, Herbold CW, Rivera MC, Servin JA, Skophammer RG.. 2007. Rooting the tree of life using nonubiquitous genes. Mol Biol Evol. 24(1):130–136. [DOI] [PubMed] [Google Scholar]
- Lake JA, Skophammer RG, Herbold CW, Servin JA.. 2009. Genome beginnings: rooting the tree of life. Philos Trans R Soc Lond B Biol Sci. 364(1527):2177–2185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merhej V, Notredame C, Royer-Carenzi M, Pontarotti P, Raoult D.. 2011. The rhizome of life: the sympatric Rickettsia felis paradigm demonstrates the random transfer of DNA sequences. Mol Biol Evol. 28(11):3213–3223. [DOI] [PubMed] [Google Scholar]
- Nelson-Sathi S, et al. 2015. Origins of major archaeal clades correspond to gene acquisitions from bacteria. Nature 517(7532):77–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pace NR. 2006. Time for a change. Nature 441(7091):289.. [DOI] [PubMed] [Google Scholar]
- Raoult D. 2010a. The post-Darwinist rhizome of life. Lancet 375(9709):104–105. [DOI] [PubMed] [Google Scholar]
- Raoult D. 2010b. Dépasser Darwin. Paris, France: Edition Plon. [Google Scholar]
- Raoult D. 2013. Of ignorance and blindness. Ebook Kindle, Amazon.
- Raoult D, Koonin EV.. 2012. Microbial genomics challenge Darwin. Front Cell Infect Microbiol. 2:127.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rivera MC, Lake JA.. 2004. The ring of life provides evidence for a genome fusion origin of eukaryotes. Nature 431(7005):152–155. [DOI] [PubMed] [Google Scholar]
- Rokas A, Williams BL, King N, Carroll SB.. 2003. Genome-scale approaches to resolving incongruence in molecular phylogenies. Nature 425(6960):798–804. [DOI] [PubMed] [Google Scholar]
- Spang A, et al. 2015. Complex archaea that bridge the gap between prokaryotes and eukaryotes. Nature 521(7551):173–179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Viezens J, Arvand M.. 2008. Simultaneous presence of two different copies of the 16S rRNA gene in Bartonella henselae. Microbiology 154(Pt 9):2881–2886. [DOI] [PubMed] [Google Scholar]
- Woese C, Fox G.. 1977. Phylogenetic structure of the prokaryotic domain: the primary kingdoms. Proc Natl Acad Sci U S A. 74(11):5088–5090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woese CR. 1987. Bacterial evolution. Microbiol Rev. 51(2):221–271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woese CR, Kandler O, Wheelis ML.. 1990. Towards a natural system of organisms: proposal for the domains Archaea, Bacteria, and Eucarya. Proc Natl Acad Sci U S A. 87(12):4576–4579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolf YI, Rogozin IB, Grishin NV, Koonin EV.. 2002. Genome trees and the tree of life. Trends Genet. 18(9):472–479. [DOI] [PubMed] [Google Scholar]
- Yutin N, Makarova KS, Mekhedov SL, Wolf YI, Koonin EV.. 2008. The deep archaeal roots of eukaryotes. Mol Biol Evol. 25(8):1619–1630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhaxybayeva O, Gogarten JP.. 2004. Cladogenesis, coalescence and the evolution of the three domains of life. Trends Genet. 20(4):182–187. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.