Figure 3.
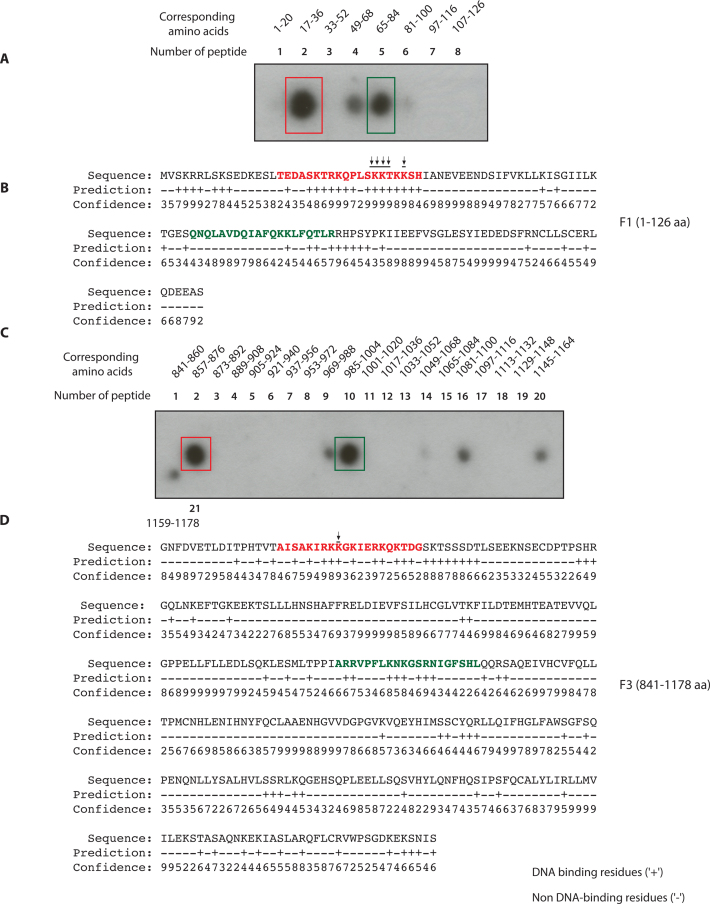
Identification of FANCD2 DNA binding residues by synthetic peptide spot assays and bioinformatic analyses (A) The synthetic peptides of 20 aa length with overlap of four amino acids (aa) were generated corresponding to F1 fragment aa 1–126. These peptides were spotted on a nitrocellulose membrane and tested for binding with radiolabeled single-strand DNA. (B) BindN prediction of highly probable DNA binding residues in F1 (high score hits are in red and lower score in green). (C) The aforementioned synthetic peptide approach was adapted for F3. The number of peptides and the corresponding amino acid number are indicated. It was seen that F3 had two intense regions corresponding to second peptide (861–876 aa) and tenth peptide (985–1004 aa). (D) Hit map of the highly probable and predicted DNA binding residues in F3 domain by BindN analysis. Peptide 2 (marked by red color) and peptide 10 (marked by green color) are shown.