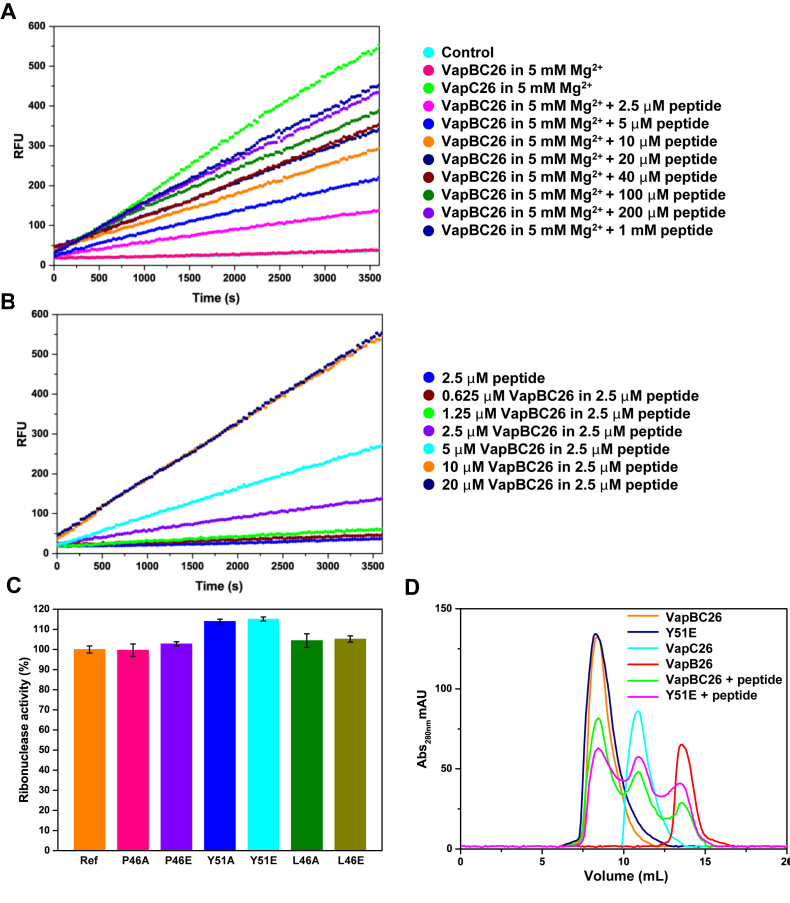
Figure 9.
Ribonuclease activity of Mycobacterium tuberculosis VapC26 measured using a VapC26 α4-mimicking peptide and experiments using mutants. (A and B) The pretreatment was performed as described in Figure 8, and 5 mM Mg2+ was added to each well for optimal activity. (A) The concentration of VapBC26 was fixed at 2.5 μM, and the concentration of peptide was increased from 2.5 μM to 1 mM. The peptide inhibited the binding of VapB26 to VapC26 by ∼50% at 10 μM and by 80% at 200 μM. (B) The concentration of the α4-mimicking peptide was fixed at 2.5 μM, and the concentration of VapBC26 was increased from 0.625 to 20 μM. Based on the data, 2.5 μM peptide interacts with ∼10 μM VapBC26. The experiment was performed in triplicate. (C and D) Comparison of the ribonuclease activity of native VapBC26 and mutated VapBC26. Pretreatment was performed as described in panels A and B. (C) A total of 10 μM of the α4-mimicking peptide was added to the mutated complex (P46A, P46E, Y51A, Y51E, L46A and L46E). Native VapBC26, which shows approximately half the activity of VapC26 in the presence of 10 μM of the α4-mimicking peptide, was used as a reference for comparison. The concentration of each VapBC26 mutant 2.5 μM was the same as that of the native VapBC26 complex. RFU obtained with the reference was taken as 100%. Tyr51 of VapB26 showed the largest effect on the binding of VapB26 and VapC26. Error bars represent the standard deviation of three replicate reactions. (D) Size exclusion chromatography of various proteins. The UV absorption at 280 nm is plotted as a function of the elution volume.