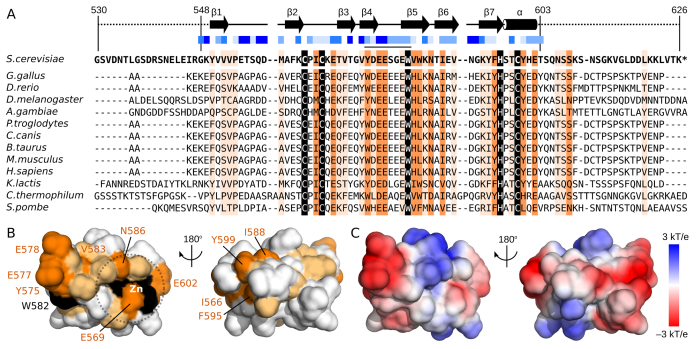
Figure 5.
Surface residue conservation separate from the zinc-binding site. (A) Sequence alignment of the second zinc-binding domain of Pcf11 from 13 species. Boundaries of the construct used in NMR studies (residues 530–626) and of the calculated structure (residues 548–603) are indicated, as are the locations of β-strands (arrows) and the α-helix (cylinder). Solvent accessibility for sidechains are calculated by using NACCESS (52) and indicated with varying shades of blue squares from solid blue (>80% accessible) to white (<20% accessible). Residues with complete sequence identity or similarity is highlighted in black and orange, respectively. Residues with similarity to yeast Pcf11 in >75% of the sequences are highlighted in light orange. (B) Surface representation of Pcf11 (548–603) with residues coloured by conservation as in panel A. The conserved patch formed by residues Y575, E577, E578 and W582 as well as the conserved residues around the Zn2+ atom (E569, V583, N586, E602) are indicated. (C) Surface charge properties of Pcf11 (548-603) with the negatively charge acidic regions in red, and the positively charged basic regions in blue.