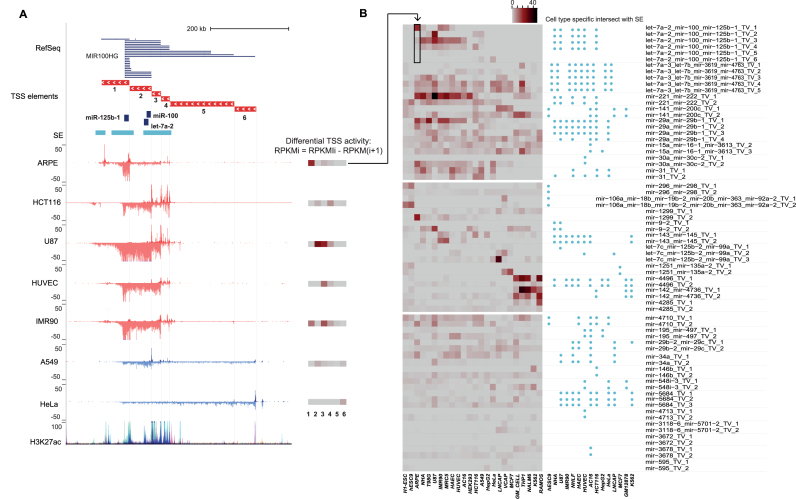
Figure 3.
Quantification of TSS elements that are used for cell type specifically. (A) The use of cell type-specific TSS can be quantified based on the TSS-elements (see ‘Materials and Methods’ section), as exemplified at the hsa-mir-100∼let-7a-2∼mir-125b-1 locus (chr11:121 723 037–122 387 271) that harbors six differentially active TSS, as supported by the Refseq annotation track. The differential TSS activity calculated based on the GRO-seq signal difference between adjacent elements (see equation in the legend and ‘Materials and Methods’ section) is summarized as a heatmap next to each track, where darker color tones correspond to higher TSS activity (in RPKM). The TSS numbering increases with the distance from the pre-miRNA. Super-enhancer locations indicated above the GRO-seq tracks can be compared to the ENCODE H3K27ac track (both correspond to overlaid activity from multiple cell types). (B) Cross-lineage quantification of differential TSS activity (left) and proximity to cell type-specific SE (right, ±100 kb) at intergenic pri-miRNA loci with multiple TSS is shown. The transposed heatmap panel corresponding to the ARPE data shown in (A) is indicated by an arrow. The heatmap is organized into three sections: the upper part of the heatmap corresponds to miRNA loci with highly dynamic TSS activity across multiple cell types; the middle part shows miRNA loci with cell-specific high activity; the lower part corresponds to miRNAs with moderate to low expression level across cell types. The ordering of the cell types reflects their origin: H1-ESC (stem cell); the neuronal lineage cell types ARPE, NHA (normal), T98G and U87 (cancer); mesoendodermal cell types IMR90, MRC5 (fibroblast), HAEC, HUVEC (endothelial), AC16 (cardiomyocyte) and HEK293T (kidney); cancer cell lines HCT116, A549, HEPG2, HeLa, LNCaP, VCAP, MCF7; blood lineage cell types (GM lymphoblastoid, normal), THP1, Nalm6, K562 and Ramos (cancer).