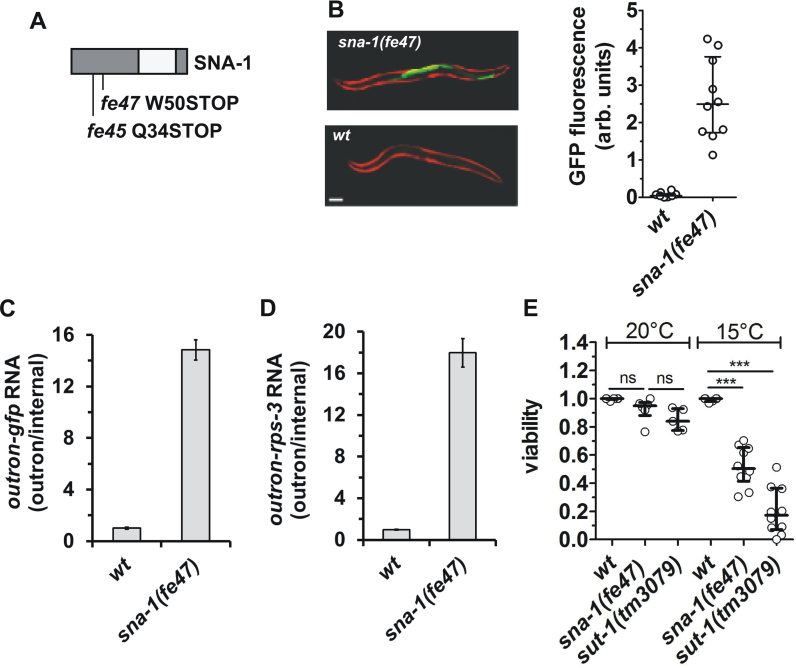
Figure 2.
Loss-of-function mutations in sna-1 recovered from a mutagenesis screen for genes involved in SL1 trans-splicing. (A) Schematic representations of the location of the residues affected by the mutations in the SNA-1 protein. The regions in dark gray are conserved in nematode SNA-1 orthologues (Supplementary Figure S1). (B) Representative images of wild type (wt) and sna-1(fe47) animals transgenic for Pvit-2::outron::gfpM1A, showing intestinal GFP (green) expression and constitutive mCherry (red) expression in body wall muscle cells. Scale bar represents 100 μm. The graph plots the GFP fluorescence of 10 animals analyzed; shown are the median and the first and third quartile. (C and D) RT-qPCR analysis of SL1 trans-splicing in wt and sna-1(fe47) animals carrying the Pvit-2::outron::gfpM1A transgene. Trans-splicing of gfp reporter gene (C) and rps-3 transcripts (D) was analyzed as described in the legend of Figure 1, with levels in wt animals set as 1. Error bars indicate standard deviation from three technical replicates. (E) Viability of wt, sna-1(fe47) and sut-1(tm3079) animals at 20 and 15°C, as expressed by the proportion of eggs that develop to L4/adult stage in 5–10 independent experiments. (***P ≤ 0.001; ‘ns’ not significant; ANOVA). Note that while at 20°C there is a slight reduction in viability in sut-1(tm3079) animals, this is not significantly different from the viability of sna-1(fe47) animals.