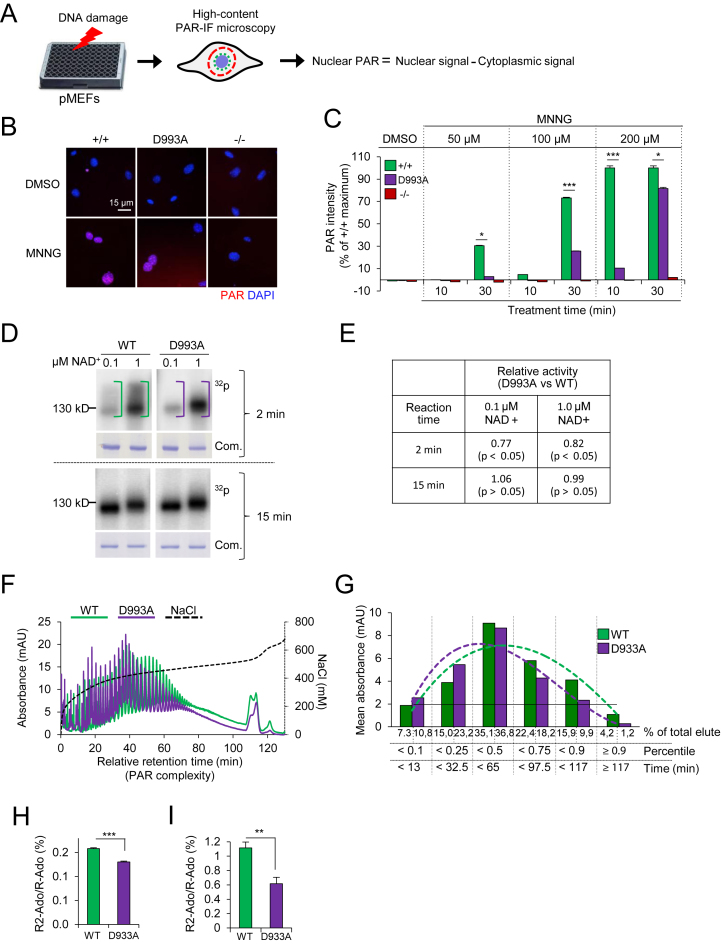
Figure 1.
PARP1D993A mutation slows enzyme kinetics of PARP1 and impairs PAR complexity. (A) Scheme of a PAR formation assay by immunofluorescence (IF) staining of PAR (PAR-IF) and data analysis. (B) Representative pictures of the PAR-IF staining of pMEFs with the indicated PARP1 genotypes (wild type PARP1 (+/+); PARP1D993A/D993A (D993A) and PARP1 knockout (–/–)) after treatment with 200 μM MNNG or DMSO for 30 min. (C) Quantification of the PAR intensity in individual cells treated for 10 min or 30 min with the indicated doses of MNNG. The DMSO controls were analysed at both time points and only 30 min is shown. The data are the means ± SEM from at least 500 cells per condition normalized against the maximum wild type (WT) level. Similar results were obtained in three independent experiments using four pMEFs littermate pairs. Asterisks (*) indicate the difference of PARP1D993A/D993A versus WT (+/+); *P <0.05; **P <0.01; ***P <0.001, as determined by a two-way ANOVA with a Tukey′s post-test. (D) In vitro activity assay of recombinant WT and D933A mutant PARP1 proteins incubated for either 2 min or 5 min with oligonucleotides mimicking DNA strand breaks and the indicated concentrations of NAD+ containing 150 nM 32P-labeled NAD+. At each reaction time, an autoradiography (32P) and Coomassie staining of the gel (Com.) are shown. (E) Quantification of the 32P signals of the panel (d) after 2 min or 15 min of reaction time at the indicated concentrations of NAD+ expressed as ratio of D993A to WT. Data are the means ± SEM from three independent experiments. P-values were calculated by the Student's t-test. (F) Analysis of the PAR chain complexity by HPLC. HPLC-DAD chromatograms of in vitro synthesized PAR from recombinant PARP1 proteins (WT and D993A mutant). The individual peaks indicate PAR molecules of different chain lengths. The retention time at 5 min was set as the starting point (‘0 min’). The complexity of PAR increases with the relative retention time. (G) The retention times from the HPLC chromatograms in (F) are grouped based on their percentiles. The resulting thresholds in min (X-axis) are plotted against the respective averaged absorbance (‘mean absorbance’ on the Y-axis). Note that the dotted trend curve of PARP1D993A is shifted to the left, indicative of less PAR complexity. (H) and (I). UPLC–MS/MS analysis of branching levels of in vitro synthesized PAR from recombinant human PARP1 (WT and D993A mutant) (H) and PAR isolated from immortalized MEFs of the indicated genotype (I). The ratio of signal intensities from the digestion products di-ribosyladenosine (R2-Ado), which is specific for PAR branching points, and ribosyl-adenosine (R-ado), which is specific for the linear part of PAR, were used to analyse the degree of PAR branching. Data are the means ± SEM of three biological (H) and five technical replicates (I). The significance is determined by Student's t-test. **P < 0.01; ***P < 0.001.