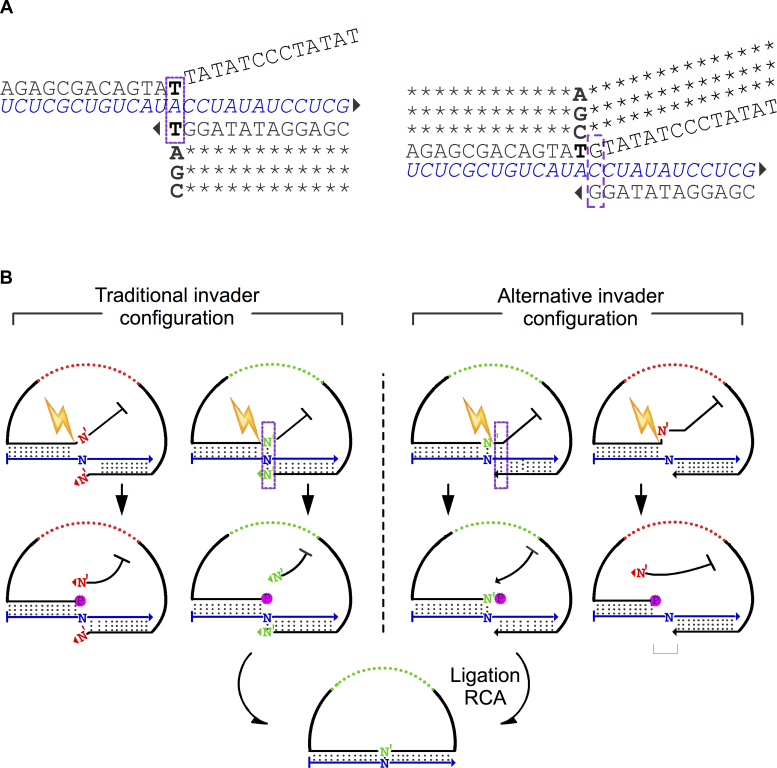
Figure 3.
iLock activation using the invader principle. (A) The structures of iLock RNA binding modes according to traditional and alternative invader configurations. In a traditional configuration, the terminal 3′ iLock base displaces the first base in the flap from the invasive junction, and competes for hybridization to the SNP position. Alternatively, the discriminatory base is localized in the 5′ iLock arm, at a single base distance from the traditional invasive junction. Bold: SNP discriminatory bases; bases participating in an invader structure formation are highlighted with a purple rectangle; triangle: 3′ OH group. (B) The invader structure is formed if a matching iLock hybridizes with a template (iLocks with a green backbone). When Taq activates (spark) matching hybridized iLock in the traditional hybridization configuration, the flap is displaced, 5′P is exposed and a discriminatory base (N’) is positioned in the 3′ iLock terminus. In the alternative invader structure configuration, complementarity between iLock 5′ arm and RNA target allows for an/the (formation of) invader structure formation. After activation, phosphorylated discriminatory base (N’) is localized on the iLock 5′ terminus. When a mismatching iLock is annealed to the target, invader structure formation and iLock activation are compromised. A combination of an imperfect 3′ arm invasion (traditional approach) and a lack of 5′ arm complementarity renders the iLock probe non-functional. Misaligned iLock in the alternative approach might generate a single nucleotide gap or a 5′ mismatch, upon activation.