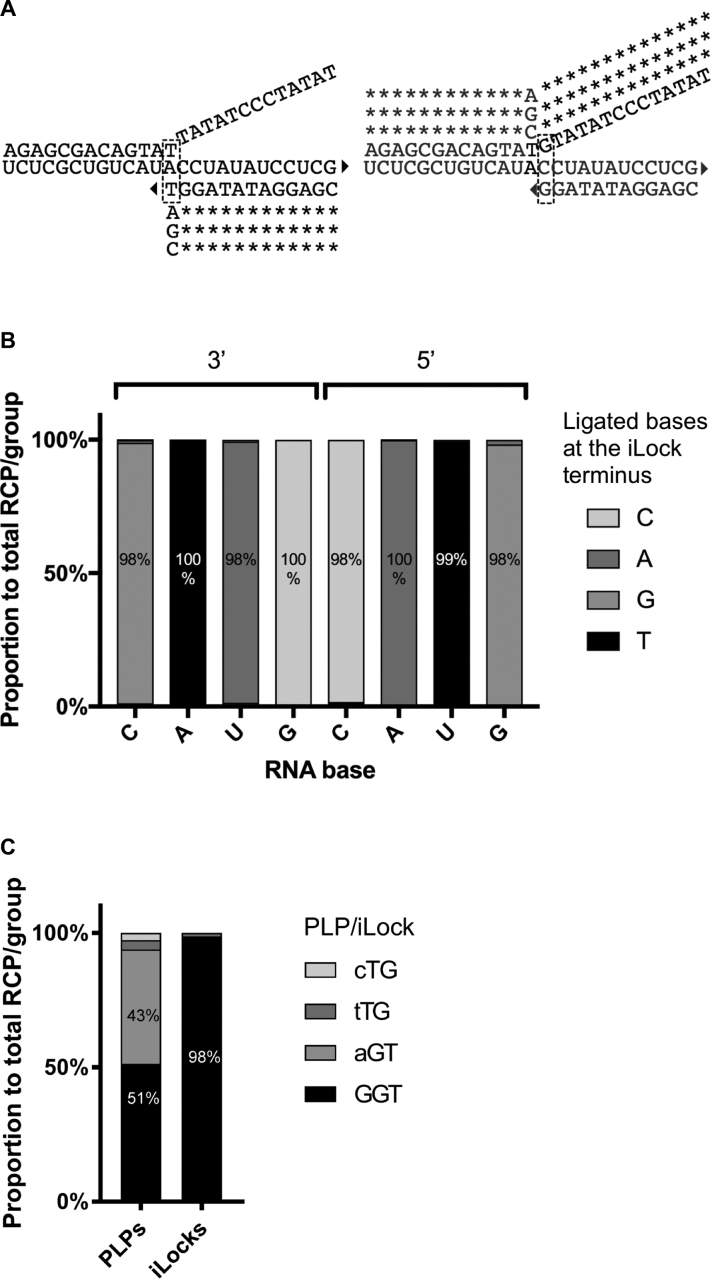
Figure 5.
Effects of 3′ and 5′ base mismatches on RNA templated DNA end-joining fidelity of PBCV-1 DNA using iLocks. (A) Probe alignment schematics using an RNA template with base A in the central, polymorphic position. Triangle: 3′ OH group; dashed rectangle: bases participating in the invader structure formation. (B) In each iLock group, average number of RCPs was added and ligation fidelity for each iLock was is presented as a percentage within the group. (C) Wild-type KRAS codon 12 (GGT) was genotyped with a complementary and three mismatching (depicted as a lowercase SNP nucleotides in the figure legend) PLPs and iLocks. Average number of RCPs in negative controls was subtracted from all the RCPs counts.