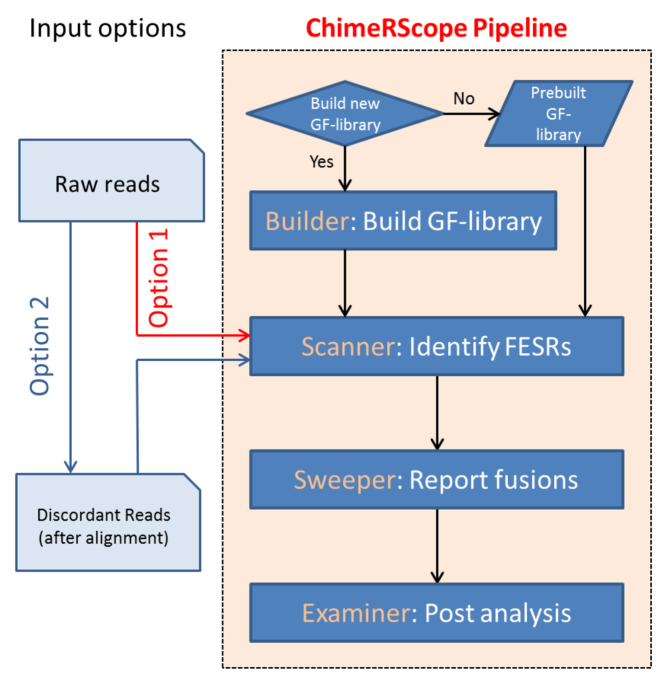
Figure 2.
The flowchart of the ChimeRScope pipeline for fusion transcript prediction. ChimeRScope can be used completely independently (Option 1) or as an add-on module in the standard RNA-Seq analysis pipeline by using the unmapped reads as input (Option 2) from third-party alignment tools. Four different modules are integrated into the ChimeRScope repository. ChimeRScope builder is a one-time step for GF-library creation and it is often carried out before the analysis. ChimeRScope Scanner identifies FESRs from input reads by using the k-mer library generated by ChimeRScope Builder. ChimeRScope Sweeper then summarizes the FESRs and outputs the list of identified fusion transcripts. Lastly, ChimeRScope Examiner will analyze fusion transcript pairs and the corresponding FESRs for fusion transcript junctions with graphical representations.