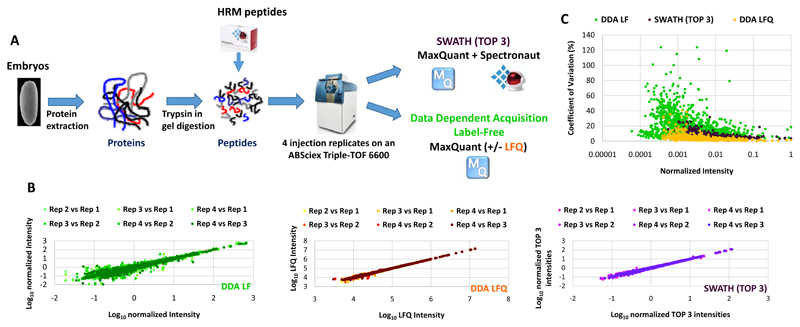
Figure 1. Comparison of Label-Free quantification methods.
A) Overall strategy for comparing SWATH and Data Dependent Acquisition Label-Free quantification methods. Protein were extracted from a Drosophila embryo sample, digested with trypsin and retention time normalisation (HRM) peptides spiked into the peptide sample before injection. Four analytical replicates were performed for SWATH and DDA acquisition modes on an ABSciex Triple-TOF 6600. The resulting files were analysed with MaxQuant with (DDA LFQ) or without (DDA LF) the LFQ module enabled or with MaxQuant and Spectronaut for the DDA and SWATH analysis, respectively. B) The Log10 intensities of the injection replicates were plotted pairwise to evaluate the run to run reproducibility of each quantification method. C) The Coefficient of variation (CV) was calculated for each protein quantified using the different quantification methods. The CVs were plotted as function of the intensities normalized to the maximal intensity.