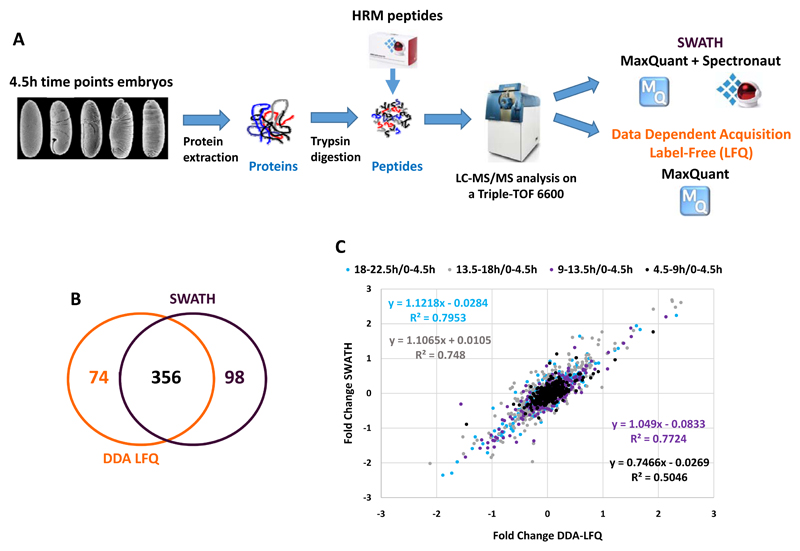
Figure 3. Monitoring the dynamics of the proteome during embryogenesis by a combination of SWATH and DDA Label-Free quantification approaches.
A) Overall strategy to measure changes in protein expression levels across D. melanogaster embryonic development by SWATH and Data Dependent Acquisition Label-Free quantification methods. Embryos were collected for a developmental time course of five 4.5 hour timepoints. Proteins were extracted, digested with trypsin and HRM peptides spiked into the peptide sample before injection. The samples were analysed using both SWATH and DDA acquisition modes on an ABSciex Triple-TOF 6600 equipped with a microflow source. The resulting files were analysed with MaxQuant with the LFQ module (DDA LFQ) enabled or with MaxQuant and Spectronaut for the DDA and SWATH analysis, respectively. B) Venn diagram representing the overlap between the proteins identified in our DDA LFQ and SWATH analysis. C) Correlation between the ratio obtained with the DDA LFQ and SWATH analysis. The ratios measured by DDA LFQ and SWATH (4.5-9h/0-4.5h, 9-13.5h/0-4.5h, 13.5-18h/0-4.5h and 18-22.5h/0-4.5h) are plotted for each protein. The values of the fitted linear regression are reported on the graph.