Abstract
Lateralized behavior (“handedness”) is unusual, but consistently found across diverse animal lineages, including humans. It is thought to reflect brain anatomical and/or functional asymmetries, but its neuro-molecular mechanisms remain largely unknown. Lake Tanganyika scale-eating cichlid fish, Perissodus microlepis show pronounced asymmetry in their jaw morphology as well as handedness in feeding behavior—biting scales preferentially only from one or the other side of their victims. This makes them an ideal model in which to investigate potential laterality in neuroanatomy and transcription in the brain in relation to behavioral handedness. After determining behavioral handedness in P. microlepis (preferred attack side), we estimated the volume of the hemispheres of brain regions and captured their gene expression profiles. Our analyses revealed that the degree of behavioral handedness is mirrored at the level of neuroanatomical asymmetry, particularly in the tectum opticum. Transcriptome analyses showed that different brain regions (tectum opticum, telencephalon, hypothalamus, and cerebellum) display distinct expression patterns, potentially reflecting their developmental interrelationships. For numerous genes in each brain region, their extent of expression differences between hemispheres was found to be correlated with the degree of behavioral lateralization. Interestingly, the tectum opticum and telencephalon showed divergent biases on the direction of up- or down-regulation of the laterality candidate genes (e.g., grm2) in the hemispheres, highlighting the connection of handedness with gene expression profiles and the different roles of these brain regions. Hence, handedness in predation behavior may be caused by asymmetric size of brain hemispheres and also by lateralized gene expressions in the brain.
Keywords: behavioral genetics/genomics, left-right asymmetry, neural structures, Perissodus microlepis, tectum opticum, telencephalon
Introduction
Behavioral handedness (or behavioral lateralization) is the tendency of an individual to favor one side of the body over the other. Humans are behaviorally lateralized: most of us are right-handed (89%) and a minority are left-handed (8%), whereas truly ambidextrous individuals are rare (3%) (Vuoksimaa et al. 2009). A number of studies provided evidence that not only humans, but also other vertebrates such as chimpanzees, dolphins, birds, reptiles, amphibians, and fishes, but also invertebrates do exhibit lateralized behaviors (e.g., reviewed in Vallortigara et al. 2011). For example, a preferred usage of the right hand was also observed in our closest relatives, the chimpanzees, Pan troglodytes (Hopkins and Cantalupo 2005). Right-eye preference for swimming counter-clockwise in bottlenose dolphins, Tursiops truncatus has recently been reported (Clark and Kuczaj II 2016). Behavioral biases are also documented for some birds such as parrots and cockatoos that preferentially use one foot over the other to handle food and objects more effectively (Harris 1989). Even in a spider (Scytodes globula) a bias in leg use during prey handling has been described (Ades and Ramires 2002). Although behavioral handedness is found across a variety of distantly related animal lineages, it is not clear how widespread it is. Nevertheless, it has been suggested that handed behavior may provide ecological benefits (Vallortigara and Rogers 2005), and it has an ancient evolutionary origin (Vallortigara et al. 1999).
Behavioral lateralization has been suggested to be associated with brain asymmetry involving neuroanatomical (structural) asymmetries and/or lateralized brain functions (Rogers and Andrew 2002). The best documented case of a correlation between behavioral lateralization and brain structural asymmetries comes from humans, where the central sulcus surface on the cerebrum in the brain was found to be greater in the left hemisphere than in the right (White et al. 1994). This human structural (cortical) asymmetry has been suggested to have a molecular genetic basis (Sun et al. 2005; Johnson et al. 2009; Hibar et al. 2015). In birds such as chickens and pigeons, differential light exposure to one eye during development gives rises to a structural asymmetry in the visual pathways that is driven by lateralized behavior (Rogers and Deng 1999). The relationship between behavioral lateralization and neural structure asymmetry has also been shown in other vertebrates such as zebrafish (Concha et al. 2000) and the convict cichlid fish, Amatitlania nigrofasciata (e.g., in the habenular nucleus; Gutiérrez-Ibáñez et al. 2011). However, examples of such a correlation, and possible cause-effect relationship, remain few (Gutiérrez-Ibáñez et al. 2011), and the functional basis of this association is still only poorly understood (Ichijo et al. 2017).
Evidence for the functional brain asymmetries related to behavioral lateralization is relatively well documented in humans, for example, functional hemispheric specialization on language and other cognitive functions predominantly localized in the left half of the brain, whereas spatial recognition is localized in the right (Galaburda et al. 1978; Gazzaniga 2005; Sun and Walsh 2006). Indeed, most right-handed humans show a strong language specialization towards the left brain hemisphere, whereas left-handed individuals indicate less distinct patterns (Coren 1992). Several studies of other vertebrates have further demonstrated that their two brain hemispheres also have distinctive functional roles (reviewed in MacNeilage et al. 2009). Although ordinary (naturally occurring) behaviors such as foraging behavior were shown to be retained towards the left hemisphere in vertebrate animals examined so far, including birds, fishes, toads, baboons, and whales, wariness (e.g., in the presence of predators) or memory-based individual recognition behaviors were primarily governed by the right hemisphere (MacNeilage et al. 2009). Examples for causal relationships of laterality underlying macroscopic structural differences or size, that is, “molecular genetic associations” between lateralized behavior and differences between the brain’s hemispheres have so far been documented exclusively for humans (e.g., Sun et al. 2005; Johnson et al. 2009; but see deCarvalho et al. 2014). Future study of other vertebrate species will help to advance our understanding of the neuro-molecular basis of lateralized behavior more generally.
A well-documented case of behavioral lateralization in fish is scale-eating (lepidophagous) in cichlids of the species, Perissodus microlepis that are endemic to Lake Tanganyika in Africa (Hori 1993; Futuyma 2009). Some individuals of this species preferentially attack the left flanks of their prey fish to bite off scales (“right-handed”), others attack the right flanks (“left-handed”), and a small minority attack both flanks with similar frequencies (no “handedness”) (Lee et al. 2012). This handed behavior is already expressed early in juvenile P. microlepis at an age of 2 months (Lee et al. 2012), and is known to be correlated with morphological mouth asymmetry (fig. 1A and B; Hori 1993; Lee et al. 2010; Van Dooren et al. 2010; Kusche et al. 2012; Takeuchi et al. 2012, 2016), which is found to be under polygenic control (Lee et al. 2015; Raffini et al. 2016). This predicted relationship between morphological and behavioral laterality is sometimes less strong for laboratory-raised fish without prior experience feeding on scales from prey fish (Lee et al. 2012). The molecular basis or neuro-molecular mechanisms underlying this remarkable behavioral laterality remain unknown and are the focus of this study.
Genome-wide RNA sequencing (RNA-Seq) permits the exploration of the functional basis of the expressed genes relatively quickly and effectively (Conesa et al. 2016). In particular, RNA-Seq facilitates the study of the gene regulatory networks of ecologically or evolutionarily intriguing traits in nonmodel organisms, even if their genomes have not been sequenced yet (e.g., Elmer et al. 2010). Several recent studies using RNA-Seq have been performed to study gene expression patterns and genetic pathways underlying key ecological traits in fishes (e.g., Gunter et al. 2013; Henning et al. 2013; Manousaki et al. 2013; Kang et al. 2015). Yet, the genetic underpinnings of adaptive behavioral phenotypes and the role of gene expression differences in regulating behavior remain largely unidentified (e.g., Whitfield et al. 2003; Aubin-Horth et al. 2005; Renn et al. 2008; Drew et al. 2012; Harris and Hofmann 2014).
In the present study, we investigated whether lateralized foraging behavior in the Lake Tanganyika scale-eating cichlid fish, Perissodus microlepis, is correlated with brain neuroanatomical (structural) asymmetry (fig. 1C and D). We also performed transcriptome analyses on different regions of the fish’s brains and on both left and right hemispheres in order to link behavioral handedness to gene transcriptional differences between hemispheres in P. microlepis. For this, we investigated whether genes showed a correlation between the degree of gene expression fold-change between hemispheres and that of lateralized behavior. The specific objectives of this study were 4-fold: 1) to test whether behavioral handedness in P. microlepis is related to left-right asymmetry in neural structures; 2) to explore the tissue-specific gene expression profiles regardless of an individuals’ handed feeding behaviors and hemispheric regions; 3) to investigate if the differences in gene expression in each paired bilaterally symmetrical brain region (i.e., tectum opticum, telencephalon, and hypothalamus; fig. 1D) between left and right hemispheres can be linked to the degree of lateralized behavior; and 4) to identify potential candidate genes in each brain region that might underlie behavioral laterality. The results of our study will open a novel perspective on the functional genomic basis of this remarkable case of behavioral laterality and lay the ground for future research on behavioral genetics/genomics.
Fig. 1.
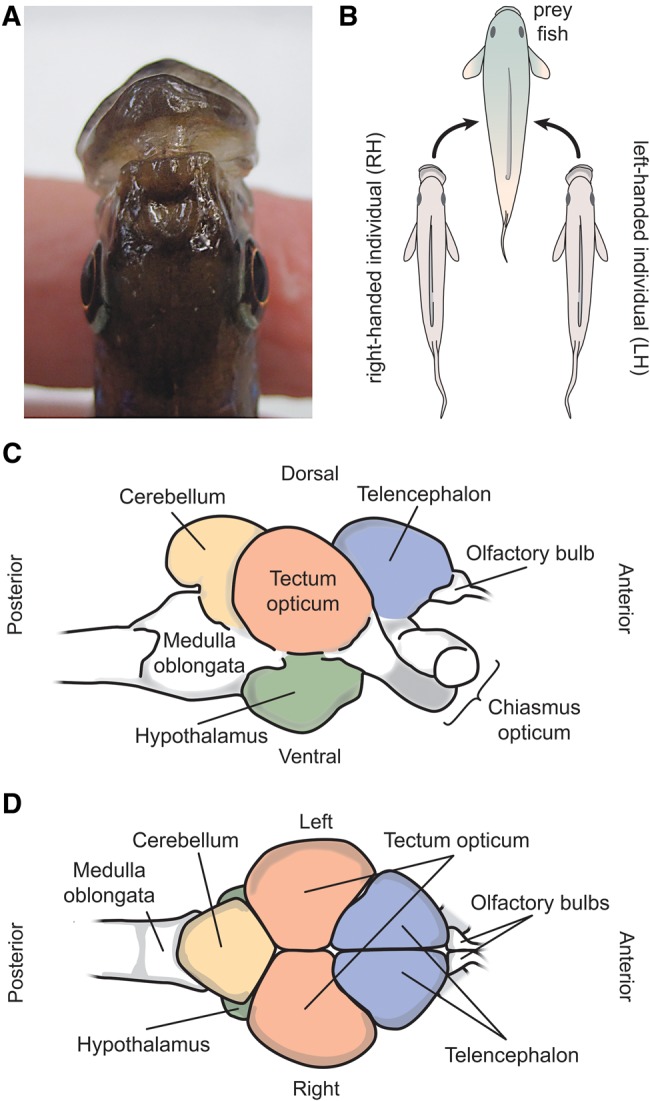
—A textbook example of a correlation between mouth asymmetry and lateralized foraging behavior in the scale-eating cichlid fish, Perissodus microlepis, from Lake Tanganyika. Note that this expected relationship is sometimes less strong in laboratory-reared fish (Lee et al. 2012; in the current study). (A) Dorsal view of left-bending mouth morph of this species. (B) Right-bending morphs preferentially attack left flanks of the prey fish (right-handed [RH]), and left-bending morphs prefer to feed from right flanks (left-handed [LH]). (C) Lateral and (D) Dorsal illustration of a Perissodus microlepis brain. Brains were dissected and the paired structures, hypothalamus, tectum opticum, and telencephalon, and the unpaired one, cerebellum, were used for gene expression analyses.
Materials and Methods
Samples
Twenty-seven Perissodus microlepis individuals were used to determine whether brain anatomical (or structural) asymmetry is correlated with behavioral foraging lateral preference. Fish were 6–10 months of age, from two laboratory-bred broods from wild-caught parents (NH × NH [no “handedness” observable, i.e. fish were showing an equal preference and attacked both flanks of prey], brood 6, N = 22; LH [“left-handed”]×NH, brood 9, N = 5) (supplementary table S1, Supplementary Material online). Twelve additional P. microlepis individuals (not included in the 27 fish) were used to carry out transcriptome analyses on the brain to investigate the architecture of gene regulatory mechanisms of behavioral lateralization (supplementary table S2, Supplementary Material online). These were also laboratory-bred fish, 6 months of age, from a single brood from wild-caught parents (RH [“right-handed”]×NH). Note that in additional 12 fish from brood 6 (NH × NH), morphological mouth asymmetry estimated from jaw-bending orientation in angle (°), after the heads of these fish were cleared and double-stained as done in Lee et al. (2015), was not associated with behavioral lateralization (Y = −0.027X + 0.207, R2 = 0.005, P = 0.836) (Lein 2012).
The parental fish for the test batches of P. microlepis were obtained in April 2010 by diving with hand nets at Toby Veal’s Lodge (S08°37.4’ E031°12’) near Mpulungu, Zambia on the southern tip of Lake Tanganyika (Kusche et al. 2012; Lee et al. 2012, 2015, 2016). The fish were kept at the animal research facility of the University of Konstanz, Germany. All the broods were reared in 40 L and later 200 L aquaria with Artemia nauplii and flake food as diet under a 12 h:12 h light:dark cycle. Animal care of the fish, foraging experiments, brain dissection, brain volume measurements and subsequent molecular experiments in the laboratory were approved by the regional board of animal welfare in Germany (Regierungspräsidium Freiburg, Abteilung Landwirtschaft, Ländlicher Raum, Veterinär und Lebensmittelwesen) (permit number: 35/9185.81/G-10/96).
Foraging Preference Experiments
To determine behavioral laterality (foraging preference = handedness) in the 39 juvenile P. microlepis, we conducted foraging experiments in a 40 L experimental tank (see Lee et al. 2012). The scale-eaters were placed individually with a single prey fish (goldfish, Carassius auratus). The test fish was placed into the trial tank at least 4 h prior to the experiment. For each fish, foraging behavior was monitored by counting the number of attacks to the left and/or right flanks on a single goldfish until a total of 20 attacks per individual were reached (with some exceptions; supplementary tables S1 and S2, Supplementary Material online) (Lee et al. 2012). Standard length (SL) of the test fish, SL of the prey fish, average time taken per attack, and our determined behavioral preferences for the 12 fish used for brain transcriptomics are shown in supplementary table S2, Supplementary Material online. There was some variation in the length of the foraging trials (total number of attacks: N = 17–21) among the 12 test fish (6.7–45.0 min; mean = 25.5 ± 12 [SD] min), which might potentially affect gene expression patterns because the time since the first attack and the collection of the brain tissue varied. The number of left and right attacks, behavioral preferences and estimated brain hemisphere volumes (see below) for the 27 fish used to examine structural asymmetry are provided in supplementary table S1, Supplementary Material online.
To determine behavioral laterality (handedness), the foraging laterality index (FLI) [FLI foraging = (number of attacks right − number of attacks left)/(number of attacks right + number of attacks left)] was calculated individually (Facchin et al. 1999). Negative values of FLI indicate more frequent attacks on the prey’s left flank (i.e., P. microlepis showed right-handed behavior), whereas positive values indicate more frequent attacks of the right flank of the prey fish (left-handed behavior; fig. 1B). For the statistical analyses, behavioral laterality was treated as a continuous variable. We also classified these fish based on the FLI into three behavioral groups, right-handed (RH) individuals, left-handed (LH) individuals, as well as individuals with no apparent handedness (NH). Fish ID 50 and 51 were considered LH fish, although binomial tests indicated marginal P values: 0.058 and 0.072 for fish ID 50 and 51, respectively (supplementary table S2, Supplementary Material online). More fish were tested for behavioral laterality, but we selected only 12 fish for the transcriptomic analysis (see below).
Brain Volume Measurements
To investigate the relationship between anatomical brain asymmetry and foraging behavior in Perissodus microlepis, brains of the 27 juvenile fish were dissected immediately after the foraging experiments and volumes of three bilaterally symmetrical brain regions (i.e., telencephalon, tectum opticum, and hypothalamus) were measured individually by the same person (E.L.) (fig. 1D), following the procedures of Pollen et al. (2007) and Gonzalez-Voyer and Kolm (2010). The test fish were euthanized by immersion in ice-slush water (2–4 °C) (Matthews and Varga 2012) right after the foraging trials and the brain volume was estimated from blind measurements with respect to the foraging type to avoid a possible observer’s bias. Olfactory bulbs were excluded from this analysis because of a relatively lower repeatability (r < 0.75) of our volume measurements detected (see below), probably due to its relatively small size (fig. 1C and D;supplementary fig. S1, Supplementary Material online). Brains were digitally photographed using a Zeiss Axiophot digital microscope (Zeiss, Germany), and width (W), length (L), and height (H) of each of the brain structures were then measured using ImageJ 1.45r (http://imagej.nih.gov/ij; last accessed October 23, 2017) to estimate their volumes (supplementary fig. S1, Supplementary Material online for measurement illustrations). According to Pollen et al. (2007) and Gonzalez-Voyer and Kolm (2010), the width (W) was measured as the greatest distance of a particular brain region that is perpendicular to the anatomical midline, the length (L) was measured as the greatest distance by that particular brain region parallel along the axis of the brain, and height (H) was measured as the greatest distance perpendicular to the body axis for that particular brain structure (supplementary fig. S1, Supplementary Material online).
The brain volume (V) was then calculated according to an ellipsoid model (Van Staaden et al. 1995): V (in mm3) = (L × W × H) π/6. The method employed in this study is a well-established and standardized protocol for the estimation of brain volumes in fishes (e.g., Pollen et al. 2007; Gonzalez-Voyer and Kolm 2010). A Spearman rank correlation analysis was conducted to test for a significant relationship between the ratio of brain hemisphere volumes (left to right) and behavioral laterality (i.e., FLI). We used a nonparametric test for this analysis because the assumption of normality was not satisfied (FLI: Shapiro–Wilk test, W = 0.897, P = 0.011). Three different Spearman rank correlation analyses were also performed for the paired brain structures.
To assess the accuracy of these measurements, repeatability (r) of the brain volume measures was estimated from independently repeated and “blind” measurements that were undertaken from two replicate photographs of each structure of the same individuals from a subsample (N = 13) using one-way ANOVA with individual as the factor, as suggested in Sokal and Rohlf (1995).
Brain Transcriptome Analyses
Brain Dissection and RNA Extraction
Brains of the experimental P. microlepis fish (N = 12) were isolated using a fine scissor and forceps in 0.1 M phosphate-buffered saline (PBS) solution on ice, immediately after the end of the foraging trials. Seven different brain regions (telencephalon-L and -R, tectum opticum-L and -R, hypothalamus-L and -R, and cerebellum) were dissected for each fish and preserved in RNAlater (Qiagen GmbH, Hilden, Germany) until RNA extraction. The tectum opticum, telencephalon, and hypothalamus are bilaterally paired symmetrical structures, whereas the cerebellum is unpaired (see fig. 1C and D;supplementary fig. S1, Supplementary Material online).
A total of 84 RNA samples from 12 fish [7 brain tissues × 4 individual fish × 3 (our determined) behavioral groups (RH, LH, and NH)] were obtained. Total RNA of each brain tissue was isolated with Trizol (Invitrogen, USA). Tissues were homogenized using pestles and mRNA was extracted with chloroform. RNA was further purified using RNeasy columns (Qiagen). On-column DNase treatment was performed following the manufacturer’s protocols. Additional washing and drying steps (washed columns twice with 80% EtOH to remove all traces of salt and ethanol) followed, and samples were spun dry for 5 min. RNA was eluted in RNase and DNase free water. RNA purity was assessed by a Nanodrop (Thermo Scientific, USA) by measuring its absorbance at 260 and 280 nm and RNA integrity (RIN) was measured using a Bioanalyzer 2100 (Agilent, USA). RNA samples with 260/280 ratio > 2.0 and RIN value > 8.5 were used for downstream library preparation.
Library Construction and Sequencing
Total RNAs recovered from the 81 brain tissues of P. microlepis (except for three RNA samples that failed qualification criteria for RNA purity or integrity: fish ID 55 hypothalamus-L and cerebellum; fish ID 57 tectum opticum-R) were subjected to a RNA-Seq protocol. Sequencing libraries were generated using the Illumina TruSeq RNA sample preparation kit (Low-Throughput protocol) according to the manufacturer’s instructions (Illumina, San Diego, CA, USA). Briefly, 500 ng of RNA was subjected to mRNA selection using poly-T oligo-attached magnetic beads followed by chemical fragmentation (5 min, 94 °C). The cleaved RNA fragments were then copied into first strand cDNA using SuperScript II reverse transcriptase (Invitrogen, USA) and Illumina proprietary random hexamer primers. After second strand synthesis using Illumina-supplied consumables, the cDNA was amplified with reagents of the same kit according to the manufacturer’s protocol and ligated to barcoded adapters. The final libraries were amplified using 15 PCR cycles. Quality assessment of the libraries was performed on a Bioanalyzer 2100 (Agilent) and the quantification was carried out in the Qubit 2.0 fluorometer (Life Technologies, USA). The 81 barcoded samples were equimolar-pooled and the same pool was loaded in 7 lanes of an Illumina flowcell in order to obtain technical replication and considerable sequencing depth. Paired-end sequencing of clustered template DNA was performed on an Illumina HiSeq2000 at the Tufts University Genomics Centre in Boston (TUCF Genomics) using four-color DNA Sequencing-By-Synthesis (SBS) technology with 210 cycles (101 cycles for each paired-read and eight cycles for the barcode sequences).
Read Quality Control
After sequencing, we obtained 1,380,519,871 raw reads of 101 bp each (average reads per sample = 17,043,455) that were quality-controlled before assembly, read mapping and downstream analyses. Reads were quality-controlled (reads with a quality score <20 were discarded) and the remaining adapters were removed with the fastx toolkit v0.013 using default parameters (http://hannonlab.cshl.edu/fastx_toolkit/; last accessed October 23, 2017). Finally, only filtered reads that were >50 nucleotides in length were used for further analyses.
Transcriptome Assembly and Annotation
For the assembly, we pooled all samples and then used a subset of the filtered reads. We chose those that were at least 95 nucleotides in length and were paired after the quality control (316,268,802 reads). 114,644,340 of those were merged and collapsed to 43,142,178 longer reads. Merging was conducted with SeqPrep (https://github.com/jstjohn/SeqPrep; last accessed October 23, 2017) and collapsing with fastx toolkit. For the assembly we used 100,812,231 nonmerged pairs of reads (total 201,624,462 reads) and 57,322,170 merged pairs of longer reads (114,644,340) collapsed to 43,142,178 reads.
The assembly process was implemented in Trinity v2.4.0 (Grabherr et al. 2011) and resulted in 305,646 transcripts. To further process the assembled transcripts and to exclude spurious ones, we mapped the reads to the assembled transcriptome using Bowtie v.1.0.0 (Langmead et al. 2009) through RSEM v1.24 (Li and Dewey 2011) to eliminate those transcripts with <1% of the reads assigned to the corresponding genes (as suggested in Haas et al. 2013). We further restricted transcripts <300 nucleotides in length to a compact and meaningful transcriptome of 176,911 transcripts clustered in 147,469 loci or components. Those sequences were used in a BLASTx search against the NCBI protein database nr (e-value threshold 1e−10). 70,004 transcripts and 48,968 loci had a significant hit in nr. Those that did not have a hit were afterwards used in a BLASTn search against NCBI nucleotide database nt (e-value threshold 1e−10). Finally, after retaining the sequences that had a significant hit in nr and/or nt we obtained 83,715 loci encompassing 110,331 transcripts that constituted our final assembly (N50: 798 bp).
Mapping and Expression Profiling
For mapping, we employed all high quality reads (1,023,729,409 reads, paired reads: 638,495,280 and single: 385,234,129) (supplementary table S3, Supplementary Material online). Mapping was conducted with Bowtie with RSEM software as implemented in the script run_RSEM_align_n_estimate.pl provided by Trinity. 722,315,809 reads (∼71%) were mapped to the assembly and assigned to the different isoforms. Expression values were estimated using RSEM at both transcript and gene levels. Given the lack of confidence in defining isoform in de novo assemblies, we conducted the expression profiling at the gene level. Paired and “orphaned” (i.e., only one of the two reads survived the filtering process) reads were mapped separately and then pooled, which provided the final count table for all 83,715 loci. The sequences of these loci were remapped to the Nile tilapia (Oreochromis niloticus) genome to retrieve protein IDs and to discard unmapped, ambiguous loci. 63,389 loci were successfully annotated with 19,145 different protein IDs and loci with the same protein IDs were summed up subsequently. The resulting count table of 19,145 proteins was normalized per sample by dividing each protein ID’s count by the median of all counts of that sample (supplementary table S4, Supplementary Material online). This normalization approach was chosen as pronounced differential expression between tissues was expected and other normalization approaches, such as used in DESeq2, rely on the assumption that only relatively few genes are differentially expressed (Dillies et al. 2013). As gene length was not considered for this normalization, reliable comparisons of expression levels among different genes were not feasible. Finally, gene names and gene ontology (GO) terms were retrieved for each protein ID from Ensembl. In spite of this, we will refer to these IDs with their corresponding gene names throughout the paper as gene expression was actually measured.
Analyses of Gene Expression Profiles
The transcriptional profiles of all 81 samples were investigated using a principle component analysis (PCA) to illustrate overall divergence in transcriptional patterns of the brain regions. Next, genes differentially expressed between brain regions were identified using pair-wise comparisons (“fdr”-corrected P values of Welch ANOVAs). Finally, gene ontology (GO) term composition of the differentially expressed genes (“test set”) was compared with the composition of all 19,145 genes found in all brain regions pooled (“reference set”) using Blast2GO (Conesa et al. 2005).
Differences in gene expression between the left and right hemispheres in the three paired brain regions were explored based on the PCA. Also, differentially expressed genes between the two hemispheres per brain region were determined as was done for the brain regions. To identify genes potentially underlying behavioral laterality, for each individual, paired brain region and gene, expression fold change was calculated by dividing the normalized gene expression value of the behaviorally preferred prey flank (e.g., in a fish with negative FLI preferentially attacked the left flank of the prey) by the value of the other nonpreferred side. The log2 of these values was then calculated, that is, if in a RH-fish a gene was upregulated in the left brain hemisphere, the log2 fold-change was a positive value and if it was upregulated in the right hemisphere the log2 fold-change was a negative value. To link the degree of behavioral lateralization to the fold change in gene expression, linear regression models were calculated for each brain region and each gene, using the absolute value of FLI (i.e., values from 0.1 to 0.8) as an independent variable and log2 fold-change of the gene as a dependent variable. Candidate genes were chosen by selecting models that showed a significant slope (P < 0.05). Additionally, genes with a model intercept potentially different from 0 (P < 0.1) were discarded as these would not fulfill the assumption that behaviorally no handed individuals have no differences in gene expression between the hemispheres. No further posthoc corrections were applied after these selection steps as our results suggested that obtained overall patterns are highly unlikely coincidental and remaining candidate genes are thus likely to be mostly “true positives” (see Results section). Nonetheless, the chosen approach is more liberal than some analyses like DESeq2 and thus particularly less significant genes should be interpreted with caution. GO term composition of these brain region-specific candidate genes was explored. Results of GO term analyses were reported and discussed only for “biological processes”, and those for “molecular function” and “cellular component” were also summarized. GO analyses were conducted using Blast2GO. All analyses of gene expression were conducted in R 3.3.0 (R Core Team 2016).
Results
Relationship between Brain Anatomical Asymmetry and Handed Feeding Behavior
Our measurements of brain hemisphere volumes were highly repeatable—estimated repeatability (r) was 0.88 (left [L] side) and 0.89 (right [R] side) for the telencephalon, 0.78 (L) and 0.88 (R) for the tectum opticum, and 0.91 (L) and 0.87 (R) for the hypothalamus. These estimates of the repeatability suggest that 78 − 91% of the total observed variation results from underlying “true” variation in the brain hemisphere volumes among individuals, and the remaining 9 − 22% variation is caused by measurement error.
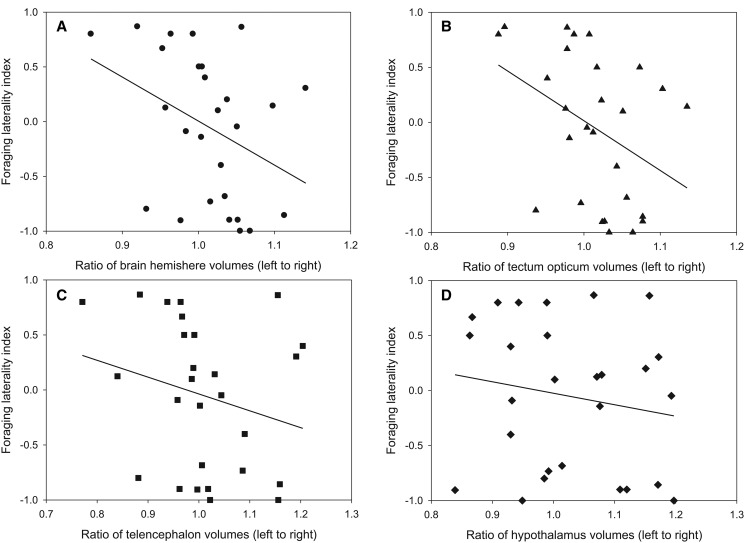
A significant negative correlation was found between the ratio of the brain hemisphere volumes and the lateralized foraging behavior [Spearmans’ rho (ρ)= −0.394, N = 27, P = 0.042; fig. 2A), which suggests that left-handed (LH) fish tend to have a larger right hemisphere than left hemisphere, whereas right-handed (RH) fish have a relatively larger left hemisphere (fig. 2A). At the brain-structure level only the tectum opticum contributed to this observed significant negative relationship (ρ = −0.455, N = 27, P = 0.017; fig. 2B). The other brain regions appear to be symmetrical (telencephalon: ρ = −0.302, N = 27, P = 0.125; fig. 2C, hypothalamus: ρ = −0.186, N = 27, P = 0.353; fig. 2D), albeit a similar trend.
Fig. 2.
—Relationship between brain anatomical left–right asymmetry and lateralized foraging behavior in P. microlepis. (A) A significant negative correlation was detected between ratio of brain hemisphere volumes (left to right) and foraging laterality index [Spearmans’ rho (ρ) = −0.394, N = 27, P = 0.042]. The brain structures used for the estimates of brain hemisphere volume include tectum opticum (B), telencephalon (C), and hypothalamus (D). At the brain-structure level, only the tectum opticum showed a significant negative relationship (ρ = −0.455, N = 27, P = 0.017).
Global Expression Patterns
To estimate the relative abundance of transcripts for each locus, we mapped the 1,023,729,409 high quality reads of the 81 samples to the assembly and assessed their expression profiles at the gene level. Global expression patterns of the 81 samples are shown in a PCA plot (fig. 3; supplementary fig. S2, Supplementary Material online). Principle components (PCs) 1–3, explaining together ∼86% of the total variation, showed pronounced differences among the four brain regions investigated (fig. 3; supplementary fig. S2, Supplementary Material online). The cerebellum’s gene expression profile appeared to be most divergent from the other three brain regions, as PC1 suggests. Although the telencephalon and hypothalamus load similarly on PCs 1 and 2, PC3 showed a clear distinction between these two regions (supplementary fig. S2, Supplementary Material online). None of the PCs accounting cumulatively for ∼95% of total variation showed any differences between the left and right hemispheres for any brain region (supplementary fig. S2, Supplementary Material online).
Fig. 3.
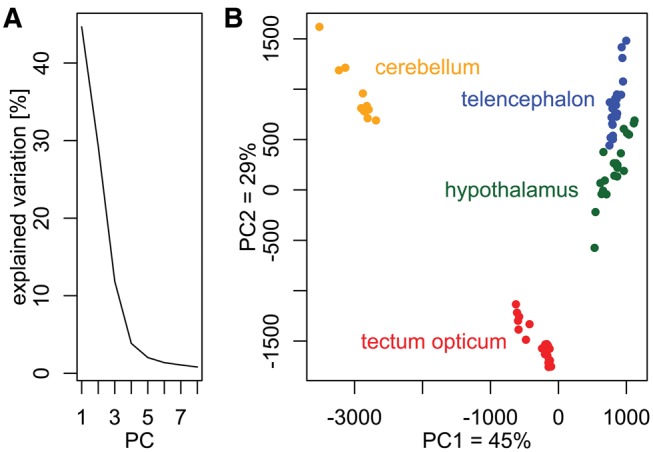
—PCA (Principal Component Analysis) of normalized count data from all 81 RNA samples. (A) A scree plot showing the percentage of explained variation per principal component (PC). The first eight PCs explained ∼95% of all variation of the dataset. (B) Scatter-plot of PC1 and PC2 (∼74% of variation). Gene expression profiles were distinct among the four brain regions, particularly the cerebellum, and the tectum opticum was different from the telencephalon and the hypothalamus, which had somewhat more similar loadings on PC1 and PC2.
A large number of genes showed differential expression in pair-wise comparisons of brain regions (left and right hemispheres pooled) after fdr corrections (table 1). Notably, approximately three quarters of the 19,145 genes expressed in the brain were differentially expressed between the cerebellum and any other brain regions, when each of the genes was tested. Comparing the hypothalamus to the telencephalon and to the tectum opticum resulted in 7,911 and 7,264 differentially expressed genes, respectively, and 11,370 genes were found to be differentially expressed between the tectum opticum and the telencephalon. In the GO term analyses we found that in the pair-wise comparisons of tectum opticum, telencephalon, and hypothalamus no highly represented GO category (in % sequences) appeared to be over- or under-represented in the differentially expressed genes compared with the overall reference gene-set (supplementary fig. S3, Supplementary Material online). Instead, most analyzed genes were represented in many GO categories (included in the “others” category). In contrast, when the tectum opticum and the hypothalamus were individually compared with the cerebellum, large percentages of the sequences were assigned to three and two GO categories, respectively, with different abundances between the candidate genes and the reference (supplementary fig. S3, Supplementary Material online). In both comparisons, genes associated with the GO category “integral component of membrane” were over-represented in the list of differentially expressed genes, whereas the category “nucleus” was under-represented, compared with the reference (supplementary fig. S3, Supplementary Material online). Finally, a very large proportion (∼98%) of the GO terms assigned to the differentially expressed genes between the telencephalon and the cerebellum were represented by the 30 illustrated categories (supplementary fig. S3, Supplementary Material online).
Table 1.
Number of Differentially Expressed Genes between Different Brain Regions Irrespective of an Individual’s Attack Behaviors and Brain Region Hemisphere
| Brain regions | Telencephalon | Hypothalamus | Cerebellum |
|---|---|---|---|
| Tectum opticum | 11,370 | 7,911 | 14,765 |
| Telencephalon | — | 7,264 | 15,133 |
| Hypothalamus | — | 14,874 |
We tested for differential gene expression between the left and right hemispheres in the three paired brain regions. Welch-ANOVA was performed if at least seven nonzero expression values were available for each hemisphere. Prior correction for multiple testing, we identified 367, 370, and 469 differentially expressed genes in the tectum opticum, telencephalon, and hypothalamus, respectively. However, after false discovery rate correction, none remained significant (all P fdr > 0.05).
Gene Expression Patterns Reflecting Behavioral Laterality
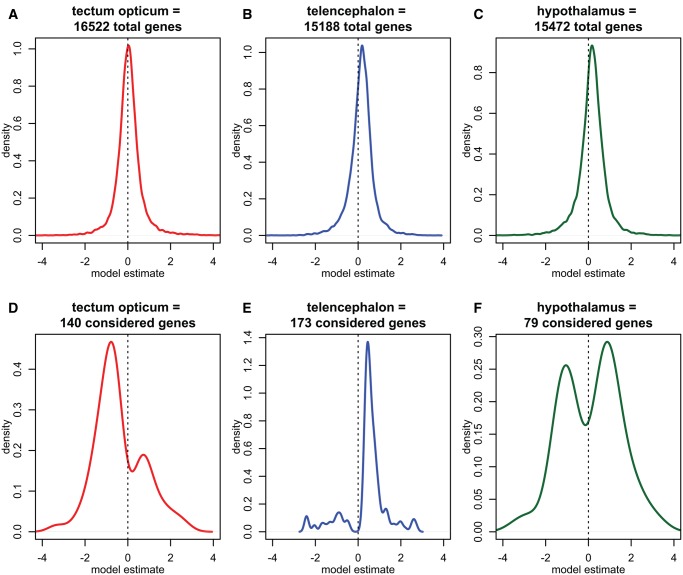
For each gene and brain region, a linear model with FLI as an independent variable and gene expression as a dependent variable was calculated (linear model slope estimates irrespective of significance are shown in fig. 4A–C). For the three paired brain regions examined, including tectum opticum, telencephalon, and hypothalamus, we identified 140, 173, and 79 candidate genes, respectively, that showed a significant linear relationship with the FLI and an intercept not different from zero (table 2; supplementary table S4, Supplementary Material online). Brain regions appeared to have different biases on the direction of candidate gene slopes, that is, whereas tectum opticum-specific candidate genes mostly had a negative slope (i.e., are up-regulated in the brain hemisphere facing the prey fish), candidate genes of the other two brain regions showed the opposite trend (figs. 4D–F and 5). This was particularly pronounced in the telencephalon, where ∼85% of candidate genes had a positive slope, that is, the genes were relatively up-regulated in the brain hemisphere that did not face the side at which prey fish were preferred (figs. 4E and 5). This unequal distribution of slope estimates suggests that identified candidate genes are mostly not false positives, which is why no post hoc correction was applied.
Fig. 4.
—Density plots of linear model slopes for the three paired brain regions. (A–C) Density plots of the linear regression model slopes (i.e., the slope estimate) of all genes tested in the respective brain region prior to candidate gene selection. (D–F) Density plots of the linear regression model slopes of selected candidate genes per brain region. (D) Most candidate genes of the tectum opticum region had a negative model slope, that is, the more behavioral laterality a fish showed, the relatively higher the gene was expressed in the brain hemisphere facing its prey. (E) In the telencephalon almost all candidate genes had a positive model slope, that is, were relatively down-regulated in the brain side facing the prey. (F) Fewest candidate genes were detected in the hypothalamus, in which slightly more genes had a positive slope than a negative slope.
Table 2.
List of the Ten Most Significant Candidate Genes in the Paired Brain Regions Including the Tectum Opticum, Telencephalon, and Hypothalamus Based on the Overall Model Fit
| EnsemblProteinID | gene | p-int. | slope | p-var. | r2 | |
|---|---|---|---|---|---|---|
| Tectum opticum | ENSONIP00000008842 | sncga | 0.7479 | −0.5608 | 0.0038 | 0.5826 |
| ENSONIP00000014770 | rtn4ip1 | 0.1936 | −1.4369 | 0.0053 | 0.5518 | |
| ENSONIP00000015841 | uhrf1 | 0.3760 | 1.7053 | 0.0065 | 0.5324 | |
| ENSONIP00000020876 | olfml2bb | 0.1367 | −1.9244 | 0.0069 | 0.5274 | |
| ENSONIP00000001535 | zgc: 110158 | 0.4366 | 0.8602 | 0.0069 | 0.5262 | |
| ENSONIP00000025694 | zgc: 64106 | 0.6089 | −1.0583 | 0.0080 | 0.5115 | |
| ENSONIP00000021527 | rbm38 (1 of many) | 0.1017 | −0.9051 | 0.0090 | 0.5001 | |
| ENSONIP00000008777 | KCNG3 (1 of many) | 0.1829 | −1.5335 | 0.0110 | 0.4787 | |
| ENSONIP00000012033 | NA | 0.1464 | −0.9119 | 0.0117 | 0.4717 | |
| ENSONIP00000018216 | zgc: 66014 | 0.4934 | −0.2808 | 0.0118 | 0.4708 | |
| Telencephalon | ENSONIP00000000966 | olfm1a | 0.1101 | 0.7254 | 0.0039 | 0.5406 |
| ENSONIP00000010660 | TOB2 | 0.2427 | −0.8450 | 0.0047 | 0.5235 | |
| ENSONIP00000002199 | cnih2 | 0.1336 | 0.5521 | 0.0074 | 0.4806 | |
| ENSONIP00000012755 | amph | 0.1016 | 0.4358 | 0.0075 | 0.4799 | |
| ENSONIP00000012999 | arhgef1a | 0.1384 | 0.3386 | 0.0082 | 0.4708 | |
| ENSONIP00000006210 | alg9 | 0.1707 | 0.6165 | 0.0083 | 0.4704 | |
| ENSONIP00000004440 | prnpb | 0.1233 | 0.3311 | 0.0086 | 0.4668 | |
| ENSONIP00000005851 | rab3b | 0.2061 | 0.6205 | 0.0109 | 0.4425 | |
| ENSONIP00000019106 | si: ch211-107o10.3 | 0.3119 | 0.5579 | 0.0127 | 0.4263 | |
| ENSONIP00000004627 | asap2a | 0.1627 | −1.0733 | 0.0141 | 0.4154 | |
| Hypothalamus | ENSONIP00000003978 | slc7a6 | 0.1117 | 1.3624 | 0.0061 | 0.5382 |
| ENSONIP00000004990 | brd1b | 0.1224 | 0.9666 | 0.0081 | 0.5102 | |
| ENSONIP00000013224 | ift20 | 0.1964 | −0.8573 | 0.0083 | 0.5086 | |
| ENSONIP00000022752 | ldlrb | 0.1469 | −1.7009 | 0.0129 | 0.4610 | |
| ENSONIP00000009502 | eps8 | 0.1512 | 1.1046 | 0.0147 | 0.4463 | |
| ENSONIP00000020992 | pappa2 | 0.1423 | 3.3755 | 0.0153 | 0.4415 | |
| ENSONIP00000003764 | dhodh | 0.1981 | 2.0406 | 0.0191 | 0.4161 | |
| ENSONIP00000009138 | mroh1 | 0.1397 | 0.6944 | 0.0194 | 0.4143 | |
| ENSONIP00000011249 | PKP4 (1 of many) | 0.1348 | 2.5339 | 0.0196 | 0.4128 | |
| ENSONIP00000002773 | pigt | 0.1201 | 0.8964 | 0.0198 | 0.4118 |
Note.—The table shows the P values of the linear model intercept, the slope estimate, the P values of the slope, the overall model P values and the model r2. Estimates for log2-fold-changes as a dependent variable.
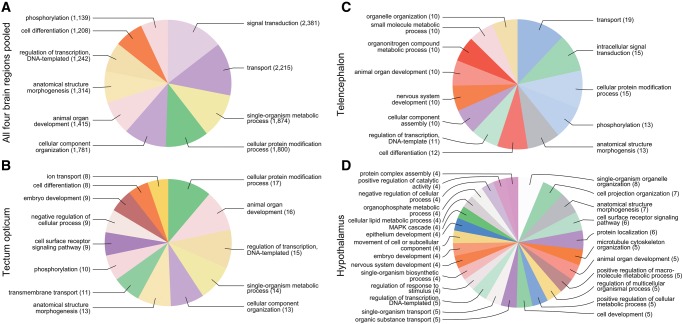
GO term analyses for biological processes suggest that laterality candidate genes in each brain region show a different composition of GO categories (fig. 6; GO term analyses for “molecular function” and “cellular component”; see supplementary fig. S4, Supplementary Material online). Notably, the laterality candidate genes of the hypothalamus were assigned to a more diverse set of GO categories (N = 26) than those of the tectum opticum (13) or the telencephalon (13) (this trend was also found if equal numbers of candidate genes per brain region were analyzed).
Fig. 6.
—Gene ontology (GO) term analysis on biological processes of all genes (A) pooled across all four brain regions, and on genes with a linear relationship to behavioral laterality in the paired (B) tectum opticum, (C) telencephalon, and (D) hypothalamus. GO term composition suggests that laterality candidate genes belong to different biological processes and their composition is brain region specific.
Discussion
The laterality (handedness) of the foraging behavior associated with an asymmetry in head morphology has made the scale-eating cichlid fish, Perissodus microlepis, from Lake Tanganyika a textbook case of the extraordinary degree of ecological adaptations (or trophic specialization) as well as for negative frequency-dependent selection (Hori 1993; Alcock 2009; Futuyma 2009) (see fig. 1A and B). However, the molecular basis or neuro-molecular mechanisms of the pronounced behavioral laterality in P. microlepis remained unexplored so far. Linking asymmetric gene expression in the brain to its functional significance, for example, lateralized behavior, has rarely been investigated and the focus of previous work remains exclusively on humans (Sun et al. 2005; Francks et al. 2007; Johnson et al. 2009; but see Nadler et al. 2006; deCarvalho et al. 2014). To our knowledge, this is the first study to link behavioral laterality to left-right asymmetry in gene expression in different brain regions as well as to neuroanatomical asymmetry, that is, the size differences between the left and right hemispheres.
Our results show that the degree of lateralized behavior in P. microlepis reflects the anatomical size differences between the left and right hemispheric regions in the brain, particularly in the visual center of the brain, the tectum opticum. This paired brain region, which belongs to the mesencephalon, is central in processing visual information and signaling stimuli to other brain regions and motor-neurons (Hubel and Wiesel 1962), involving prey-catching behavior (“orientation toward prey behavior”) as documented in salamanders (Himstedt and Roth 1980) and fishes (Al-Akel et al. 1986; Gahtan et al. 2005). It is important to note that, as was shown for other vertebrates, fish brain hemispheres typically control the opposite sides of the individual’s body, that is, the right tectum opticum processes incoming visual information from the left eye and the left tectum opticum from the right eye via the chiasmus opticum (fig. 1C and D). In fish with strong behavioral laterality, we showed that the tectum opticum hemisphere that is processing the data from the eye that is facing towards the prey during an attack is significantly larger in volume than its counterpart (fig. 2B). In fish with weak behavioral laterality this effect of neuroanatomical left-right asymmetry was not present. These findings are consistent with the patterns previously found in pigeons that the right eye is dominant over the left and accordingly the left hemisphere of their tectum opticum is increased in size (Güntürkün 1997). Our results thus indicate that although the extent of lateralization in behavior varies among individuals, its occurrence may be determined by differences in symmetry of some neuroanatomical regions. This finding also supports the notion that the neurologically more dominant side of the body may be orchestrated by the relatively larger corresponding brain hemisphere (on the contralateral side).
Genome-wide transcriptional profiles of four brain regions analyzed in this study reveal that different brain regions have distinct transcriptional profiles (fig. 3). These results agree with previous findings of regionally specific gene expression patterns in adult mice brain (Nadler et al. 2006; Lein et al. 2007) and also in adult human brain (Hawrylycz et al. 2012). Notably, the cerebellum appears to be transcriptionally the most divergent from the other three brain parts (separated from the others by PC1) as shown for adult mice brain (Nadler et al. 2006), followed by the tectum opticum (separated from the others by PC2) and finally the telencephalon and the hypothalamus appear to be transcriptionally most similar (separated from each other only by PC3). This order corresponds well to their developmental Bauplan and interrelationships, with the telencephalon (most anterior) and the hypothalamus (second most anterior) both being components of the prosencephalon (forebrain), followed by the tectum opticum in the mesencephalon (midbrain) and the cerebellum, a part of the rhombencephalon (hindbrain; figs. 1 and 5). These results are consistent with those of previous studies that found that gene expression differences across brain regions recapitulated the developmental processes of the embryonic brain in adult mice based on their cluster analyses (Zapala et al. 2005; Nadler et al. 2006). Likewise, we find a higher number of differentially expressed genes between the cerebellum and any other brain regions, and relatively fewer between the telencephalon and the hypothalamus. The GO enrichment tests on the differentially expressed genes support the transcriptionally distinct position of the cerebellum: comparisons within and between prosencephalic and mesencephalic brain regions resulted in many differentially represented GO categories with each representing only a small fraction of differentially expressed genes (supplementary fig. S3, Supplementary Material online). However, when the other brain regions are compared with the cerebellum, fewer major “biological process”-GO categories cover most differentially expressed genes, suggesting that the cerebellum is most different from the other brain regions, for example, by substantially differing from each other in genes relating to “integral components of cell membranes”, such as ion channel genes.
A central goal of this study was to determine candidate genes that relate to behavioral laterality and we identified genes potentially underpinning behavioral laterality in all three paired brain regions. For the tectum opticum, we found expression differences in 140 genes between the left and right hemispheres that could be related to behavioral lateralization (supplementary table S4, Supplementary Material online). The most significant and promising candidate is synuclein gamma (sncga) and is discussed below in more detail. Besides sncga, we identified paralogs of other putatively asymmetrically expressed neurotransmitters, such as numerous members of the solute carrier family as well as the glutamate metabotropic receptor family (their functions are summarized in deCarvalho et al. 2014; Karlebach and Francks 2015).
The function of the telencephalon and its role in behaviors in teleost fishes remain largely unstudied. The telencephalon, known as the cognitive center of the vertebrate brain, is thought to be involved in processing all sensory information especially olfactory signals coming from the olfactory bulbs, learning, directing active movements and social-reproductive behaviors (Overmier and Patten 1982; Butler and Hodos 2005). Lesion studies have suggested that the telencephalon of the teleost fish plays a role in spatial learning and memory (Kaplan and Aronson 1967; reviewed in Salas et al. 2006). We identified a pronounced bias in the slope of candidate genes relating to lateralized feeding behavior in P. microlepis, indicating that ∼85% of these genes are up-regulated in the telencephalon hemisphere away from prey fish and thus putatively controlling the body side of the scale-eater facing the prey fish. Among these telencephalic candidate genes, interestingly, we found mgrn1b that has been shown to crucially affect left-right patterning in mice and that is important for proper neuronal development (Cota et al. 2006). Lrrtm1, another candidate gene, has been suggested to be related to handedness and schizophrenia in humans (Francks et al. 2007). Changes in the expression of the telencephalic candidate kcng3, a gene coding for a potassium channel subunit, might be related to handedness, as changes in the closely related kcns3 have been suggested to be related to schizophrenia in humans (Georgiev et al. 2014). Schizophrenia is, interestingly, a condition that is associated with reduced brain lateralization and handedness (Sommer et al. 2001). Expression of two protocadherins (pcdh1b and pcdh17) was found to be significantly associated with lateralized behavior, a gene family that has been suggested to be involved with the morphological asymmetry in P. microlepis from a population genomics study (Raffini et al. 2016). Other candidates include cadm1b, cadpsb, grm2, and kctd2, which themselves or their paralogs have been reported to be expressed asymmetrically in the brain of zebrafish (deCarvalho et al. 2014). Interestingly, grm2, that had a significant negative slope in the tectum opticum, had a positive slope in the telencephalon. It thus exemplifies the overall inversion of slope biases that we observed between the two brain regions (figs. 4 and 5) that may be due to the circumstance that the tectum opticum is processing visual signals from the eye of the opposite body side while the telencephalon processes olfactory signals from the same body side.
The smallest number of candidate genes with the most subtle slope biase were detected in the hypothalamus, the brain region that lies ventrally, and developmentally between the tectum opticum and the telencephalon. Identified candidate genes include smad2, which is crucial in early embryonic development of asymmetric structures (Tremblay et al. 2000) and members of the solute carrier family (slc7a6, slc23a1, and slc33a1). In general, the hypothalamus shows the least asymmetry in gene expression in the paired brain regions that we analyzed.
Highlighting our overall evidence, we found strong support for behavioral laterality represented in the tectum opticum by its neuroanatomical and transcriptional asymmetries, as our results show a left-right size difference and also a pronounced bias in the linear model slopes in this particular brain region. The tectum opticum-specific candidate genes appear to be downregulated in the brain hemisphere that controls the body side facing the prey fish during an attack—the opposite trend we find in the telencephalon (fig. 5). Although the tectum opticum is processing signals from the eye on the opposite side of the body, the telencephalon processes information from the olfactory sensors on the same body side. These two findings combined may explain the inverted biases of gene expression patterns between these two brain regions (figs. 4 and 5). The hypothalamus is a part of the diencephalon, which is thought to be the most complex region of the teleost brain (Cerdá-Reverter et al. 2001). Among other functions, it processes information of gustatory information, vision, olfaction and reproduction (Braford and Northcutt1983; Northcutt and Wullimann 1988; Wullimann 1997). The neuroanatomical and functional diversities of the hypothalamus is probably also reflected in the most diverse set of GO terms that deviated from the reference set (fig. 6).
Fig. 5.
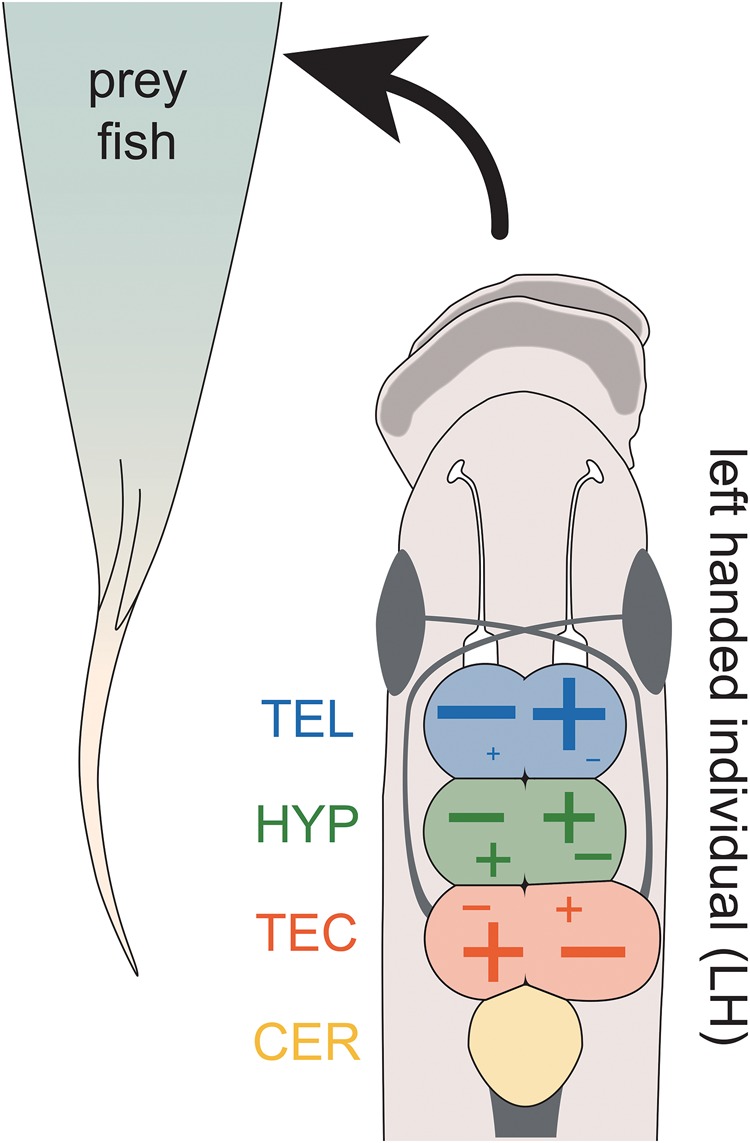
—Neuroanatomical and transcriptional patterns reflect behavioral laterality in P. microlepis. The more lateralized feeding behavior, the larger the hemisphere of the tectum opticum (TEC) that is processing the information of the body side facing the prey fish. Expression differences in the candidate genes between brain hemisphere regions were increased with increasing behavioral laterality. However, the direction of up- (+) or down- (−) regulation in candidate genes showed divergent biases among the brain regions as illustrated. In the telencephalon (TEL), ∼85% of candidate genes showed an increase in the hemisphere fold-change with an increasing feeding laterality index (FLI), that is, most genes were relatively up-regulated in the brain hemisphere not facing the prey fish. This trend was less pronounced in the hypothalamus (HYP) where only ∼60% of genes were relatively upregulated in the same hemisphere and was completely reversed in the tectum opticum (TEC) where most genes were relatively up-regulated in the hemisphere facing the prey fish.
One of the exciting findings in our study is the highly significant correlation between the degree of asymmetrical expression in sncga and that of behavioral lateralization. sncga belongs to a group of soluble proteins (synucleins) that act as neurotransmitters and are commonly expressed in neural tissues; however, sncga is typically expressed in peripheral neurons and the retina (George 2002). In zebrafish, sncga was found to be asymmetrically expressed in the habenular nuclei—a diencephalic complex that has been shown to be involved in motor behaviors and the motivation to exhibit them, and that is thought to be a hub for asymmetry in the vertebrate brain (Bianco and Wilson 2009; Chen et al. 2009; Ichijo et al. 2017). Specifically, habenular nuclei are shown to signal motor neurons and interact with the dopaminergic system, potentially leading to behavioral side preferences (reviewed in Bianco and Wilson 2009; Gutiérrez-Ibáñez et al. 2011; as suggested for P. microlepis by Ichijo et al. 2017). This is an exciting hypothesis that deserves further studies.
The causal relationships leading to the lateralized behavioral phenotypes in P. microlepis may be critically influenced by the brain anatomical and transcriptional asymmetries that we identified. Fish with structural and functional asymmetries likely controlled by a genetic component (Ichijo et al. 2017) may be stimulated to exhibit lateralized behaviors, such as feeding. Lateralized foraging behavior finally may lead to plastic changes in the oral jaw phenotype in P. microlepis (Van Dooren et al. 2010; Palmer 2012; Lee et al. 2012). Phenotypic plasticity has been shown to be a prominent feature of many cichlid fishes (e.g., Meyer 1987; Klingenberg et al. 2003; Muschick et al. 2011; Schneider and Meyer 2017) possibly as a developmental basis for both the diversity of jaw shapes and the lateralized jaws for which this species is renowned. However, previous studies seemed to show that the jaw laterality is already present in P. microlepis fry before they start actively to feed, arguing against plasticity (Hori 1993; Stewart and Albertson 2010). Our more recent studies reconciled the disputed mechanisms for morphological asymmetry where not only genes, but also environmental effects appear to be responsible for the observed variation in this trait (Lee et al. 2015; Raffini et al. 2016). To more fully resolve this issue, time-course studies investigating both morphological asymmetries in the jaw and the brain as well as differential gene expression in the brain starting at the earliest possible time-point and covering the time until clear feeding preferences (e.g., at an age of 2-months; Lee et al. 2012) would be necessary. Such studies would allow to determine the developmental sequence of asymmetry in the jaw and brain, and also behavior (Ichijo et al. 2017).
Some of our earlier work (Lee et al. 2012) and results from the current analysis suggest that behavioral laterality may occur earlier than morphological laterality in the jaw or that both may emerge independently, given only a weak correlation between mouth asymmetry and behavioral laterality in “laboratory-reared” scale-eating cichlids. We also found that in laboratory-reared juvenile fish (e.g., at an age of 2-months) that had been raised on Artemia nauplii and flake food, the degree of mouth asymmetry was relatively small based on our quantitative measurements, although we could document an early strong laterality in behavioral preference when feeding on scales (Lee et al. 2012). These findings suggest that lateralized foraging behavior might precede and “guide” morphological asymmetry via plasticity. A central question that follow-up studies should thus ask is how strongly behavioral laterality is genetically determined (and heritable). Whether the asymmetry of brain regions, particularly tectum opticum (as shown in this study) is a direct consequence of genes that cause lateralized behaviors or whether these brain regions respond to lateralized behaviors secondarily remains unclear. However, the former scenario is more plausible, given that the test fish used for brain anatomical and transcriptional asymmetries analyzed here had not had an opportunity to feed on scales from prey fish before.
Perissodus microlepis is an evolutionary text-book example of negative frequency-dependent selection due to its iconic mouth asymmetry and feeding laterality. Despite prior efforts to elucidate the genetic or environmental bases of morphological mouth asymmetry (e.g., Lee et al. 2015; Raffini et al. 2016), relatively little attention had been paid to the molecular basis of behavioral laterality so far. In this study we show that feeding laterality in P. microlepis is reflected in its neuroanatomy, particularly in the tectum opticum. Transcriptional profiles further suggest that in both the tectum opticum and telencephalon, gene expressions are also lateralized, and that these gene expression lateralities are likely to be causally linked to behavioral laterality. Given the observed neuro-molecular basis of behavioral laterality, the next work needs to focus on functional tests for the proposed causal relationships and on the heritability of the neuroanatomical and transcriptional asymmetries.
Authors’ Contributions
H.J.L. and A.M. designed the study, H.J.L. carried out fish breeding, H.J.L. and E.L. conducted foraging preference experiments and brain volume measurements. J.H.K. and P.F. performed RNA experiments, R.F.S., T.M., and P.F. analyzed and interpreted the transcriptomic data. H.J.L., R.F.S., T.M., J.H.K., P.F., and A.M. drafted the manuscript. All authors edited and agreed to a final version of the manuscript.
Supplementary Material
Supplementary data are available at Genome Biology and Evolution online.
Supplementary Material
Acknowledgments
This study was supported through grants of the Deutsche Forschungsgemeinschaft (DFG) (LE2848/1-1), the Young Scholar Fund (FP 411/12) of the University of Konstanz, and the Ministry of Oceans and Fisheries, Korea (Project title: Long-term changes in structure and function in the marine ecosystems of Korea) to H.J.L. and through support of the DFG and the University of Konstanz to A.M. We thank the University of Konstanz for supporting the Genomics Centre of the University of Konstanz (GeCKo) and the Meyer laboratory, and thank Jacques Lagnel for bioinformatics support. We also thank Francesca Raffini, Harold Zakon and Hans Hofmann for helpful comments on an earlier version of this manuscript.
Literature Cited
- Ades C, Ramires EN.. 2002. Asymmetry of leg use during prey handling in the spider Scytodes globula (Scytodidae). J Insect Behav. 15:563–570.http://dx.doi.org/10.1023/A:1016337418472 [Google Scholar]
- Al-Akel AS, Guthrie DM, Banks JR.. 1986. Motor responses to localized electrical stimulation of the tectum in the freshwater perch (Perca fluviatilis). Neuroscience 19(4):1381–1391.http://dx.doi.org/10.1016/0306-4522(86)90150-8 [DOI] [PubMed] [Google Scholar]
- Alcock J. 2009. Animal behavior. Sunderland: Sinauer Associates. [Google Scholar]
- Aubin-Horth N, Landry CR, Letcher BH, Hofmann HA.. 2005. Alternative life histories shape brain gene expression profiles in males of the same population. Proc Biol Sci. 272(1573):1655–1662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bianco IH, Wilson SW.. 2009. The habenular nuclei: a conserved asymmetric relay station in the vertebrate brain. Philos Trans R Soc Lond B Biol Sci. 364(1519):1005–1020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braford MR, Northcutt RG.. 1983. Organization of the diencephalon and pretectum of the ray-finned fishes. Fish Neurobiol. 2:117–163. [Google Scholar]
- Butler AB, Hodos W.. 2005. Comparative vertebrate neuroanatomy. New Jersey: John Wiley and Sons Inc. [Google Scholar]
- Cerdá-Reverter JM, Zanuy S, Muñoz-Cueto JA.. 2001. Cytoarchitectonic study of the brain of a perciform species, the sea bass (Dicentrarchus labrax). II. The Diencephalon. J Morphol. 247(3):229–251. [DOI] [PubMed] [Google Scholar]
- Chen Y-C, et al. 2009. Recapitulation of zebrafish sncga expression pattern and labeling the habenular complex in transgenic zebrafish using green fluorescent protein reporter gene. Dev Dynam. 238(3):746–754. [DOI] [PubMed] [Google Scholar]
- Clark FE, Kuczaj SA II.. 2016. Lateralized behavior of bottlenose dolphins using an underwater maze. Int J Comp Psychol. 29:1. [Google Scholar]
- Concha ML, Burdine RD, Russell C, Schier AF, Wilson SW.. 2000. A nodal signaling pathway regulates the laterality of neuroanatomical asymmetries in the zebrafish forebrain. Neuron 28(2):399–409. [DOI] [PubMed] [Google Scholar]
- Conesa A, Götz S, García-Gómez JM, Terol J, Talón M, Robles M.. 2005. Blast2GO: a universal tool for annotation, visualization and analysis in functional genomics research. Bioinformatics 21(18):3674–3676. [DOI] [PubMed] [Google Scholar]
- Conesa A, et al. 2016. A survey of best practices for RNA-seq data analysis. Genome Biol. 17:13.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coren S. 1992. The left-hander syndrome: The causes and consequences of left-handedness. New York: The Free Press. [Google Scholar]
- Cota CD, et al. 2006. Mice with mutations in Mahogunin ring finger-1 (Mgrn1) exhibit abnormal patterning of the left-right axis. Dev Dynam. 235(12):3438–3447. [DOI] [PubMed] [Google Scholar]
- deCarvalho TN, et al. 2014. Neurotransmitter map of the asymmetric dorsal habenular nuclei of zebrafish. Genesis 52(6):636–655.http://dx.doi.org/10.1002/dvg.22785 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dillies MA, et al. 2013. A comprehensive evaluation of normalization methods for Illumina high-throughput RNA sequencing data analysis. Brief Bioinformatics 14(6):671–683.http://dx.doi.org/10.1093/bib/bbs046 [DOI] [PubMed] [Google Scholar]
- Drew RE, et al. 2012. Brain transcriptome variation among behaviorally distinct strains of zebrafish (Danio rerio). BMC Genomics 13:323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elmer KR, et al. 2010. Rapid evolution and selection inferred from the transcriptomes of sympatric crater lake cichlid fishes. Mol Ecol. 19:197–211.http://dx.doi.org/10.1111/j.1365-294X.2009.04488.x [DOI] [PubMed] [Google Scholar]
- Facchin L, Bisazza A, Vallortigara G.. 1999. What causes lateralization of detour behavior in fish? evidence for asymmetries in eye use. Behav Brain Res. 103(2):229–234. [DOI] [PubMed] [Google Scholar]
- Francks C, et al. 2007. LRRTM1 on chromosome 2p12 is a maternally suppressed gene that is associated paternally with handedness and schizophrenia. Mol Psychiatry 12(12):1129–1139.http://dx.doi.org/10.1038/sj.mp.4002053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Futuyma DJ. 2009. Evolution. Sunderland: Sinauer Associates. [Google Scholar]
- Gahtan E, Tanger P, Baier H.. 2005. Visual prey capture in larval zebrafish is controlled by identified reticulospinal neurons downstream of the tectum. J Neurosci. 25(40):9294–9303.http://dx.doi.org/10.1523/JNEUROSCI.2678-05.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galaburda AM, LeMay M, Kemper TL, Geschwind N.. 1978. Right-left asymmetries in the brain. Science 199(4331):852–856.http://dx.doi.org/10.1126/science.341314 [DOI] [PubMed] [Google Scholar]
- Gazzaniga MS. 2005. Forty-five years of split-brain research and still going strong. Nat Rev Neurosci. 6(8):653–659.http://dx.doi.org/10.1038/nrn1723 [DOI] [PubMed] [Google Scholar]
- George JM. 2002. The synucleins. Genome Biol. 3(1):REVIEWS3002.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Georgiev D, et al. 2014. Lower gene expression for KCNS3 potassium channel subunit in parvalbumin-containing neurons in the prefrontal cortex in schizophrenia. Am J Psychiatry 171(1):62–71.http://dx.doi.org/10.1176/appi.ajp.2013.13040468 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grabherr MG, et al. 2011. Full-length transcriptome assembly from RNA-seq data without a reference genome. Nat Biotechnol. 29(7):644–652.http://dx.doi.org/10.1038/nbt.1883 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez-Voyer A, Kolm N.. 2010. Sex, ecology and the brain: evolutionary correlates of brain structure volumes in Tanganyikan cichlids. PLoS One 5(12):e14355.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gunter HM, et al. 2013. Shaping development through mechanical strain: the transcriptional basis of diet-induced phenotypic plasticity in a cichlid fish. Mol Ecol. 22(17):4516–4531.http://dx.doi.org/10.1111/mec.12417 [DOI] [PubMed] [Google Scholar]
- Güntürkün O. 1997. Morphological asymmetries of the tectum opticum in the pigeon. Exp Brain Res. 116(3):561–566. [DOI] [PubMed] [Google Scholar]
- Gutiérrez-Ibáñez C, Reddon AR, Kreuzer MB, Wylie DR, Hurd PL.. 2011. Variation in asymmetry of the habenular nucleus correlates with behavioral asymmetry in a cichlid fish. Behav Brain Res. 221(1):189–196. [DOI] [PubMed] [Google Scholar]
- Haas BJ, et al. 2013. De novo transcript sequence reconstruction from RNA-seq using the Trinity platform for reference generation and analysis. Nat Protoc. 8(8):1494–1512.http://dx.doi.org/10.1038/nprot.2013.084 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris LJ. 1989. Footedness in parrots: three centuries of research, theory, and mere surmise. Can J Psychol. 43(3):369–396.http://dx.doi.org/10.1037/h0084228 [DOI] [PubMed] [Google Scholar]
- Harris RM, Hofmann HA.. 2014. Neurogenomics of behavioral plasticity In: Landry CR, Aubin-Horth N, editors. Ecological genomics: ecology and the evolution of genes and genomics. Advances in experimental medicine and biology. Netherlands: Springer; p. 149–168. [DOI] [PubMed] [Google Scholar]
- Hawrylycz MJ, et al. 2012. An anatomically comprehensive atlas of the adult human brain transcriptome. Nature 489(7416):391–399.http://dx.doi.org/10.1038/nature11405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henning F, Jones JC, Franchini P, Meyer A.. 2013. Transcriptomics of morphological color change in polychromatic Midas cichlids. BMC Genomics 14:171.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hibar DP, et al. 2015. Common genetic variants influence human subcortical brain structures. Nature 520(7546):224–229.http://dx.doi.org/10.1038/nature14101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Himstedt W, Roth G.. 1980. Neuronal responses in the tectum opticum of Salamandra to visual prey stimuli. J Comp Physiol. 135(3):251–257. [Google Scholar]
- Hopkins WD, Cantalupo C.. 2005. Individual and setting differences in the hand preferences of chimpanzees (Pan troglodytes): a critical analysis and some alternative explanations. Laterality 10(1):65–80.http://dx.doi.org/10.1080/13576500342000301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hori M. 1993. Frequency-dependent natural-selection in the handedness of scale-eating cichlid fish. Science 260(5105):216–219.http://dx.doi.org/10.1126/science.260.5105.216 [DOI] [PubMed] [Google Scholar]
- Hubel DH, Wiesel TN.. 1962. Receptive fields, binocular interaction and functional architecture in the cat’s visual cortex. J Physiol. 160:106–154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ichijo H, Nakamura T, Kawaguchi M, Takeuchi Y.. 2017. An evolutionary hypothesis of binary opposition in functional incompatibility about habenular asymmetry in vertebrates. Front Neurosci. 10:595.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson MB, et al. 2009. Functional and evolutionary insights into human brain development through global transcriptome analysis. Neuron 62(4):494–509.http://dx.doi.org/10.1016/j.neuron.2009.03.027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang JH, et al. 2015. Transcriptomics of two evolutionary novelties: how to make a sperm-transfer organ out of an anal fin and a sexually selected “sword” out of a caudal fin. Ecol Evol. 5(4):848–864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaplan H, Aronson LR.. 1967. Effect of forebrain ablation on the performance of a conditioned avoidance response in the teleost fish, Tilapia h. macrocephala. Anim Behav. 15(4):438–448.http://dx.doi.org/10.1016/0003-3472(67)90042-5 [DOI] [PubMed] [Google Scholar]
- Karlebach G, Francks C.. 2015. Lateralization of gene expression in human language cortex. Cortex 67:30–36.http://dx.doi.org/10.1016/j.cortex.2015.03.003 [DOI] [PubMed] [Google Scholar]
- Klingenberg CP, Barluenga M, Meyer A.. 2003. Body shape variation in cichlid fishes of the Amphilophus citrinellus species complex. Biol J Linn Soc. 80(3):397–408.http://dx.doi.org/10.1046/j.1095-8312.2003.00246.x [Google Scholar]
- Kusche H, Lee HJ, Meyer A.. 2012. Mouth asymmetry in the textbook example of scale-eating cichlid fish is not a discrete dimorphism after all. Proc Biol Sci. 279(1748):4715–4723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmead B, Trapnell C, Pop M, Salzberg SL.. 2009. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 10(3):R25.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee HJ, et al. 2010. Genetic support for random mating between left and right-mouth morphs in the dimorphic scale-eating cichlid fish Perissodus microlepis from Lake Tanganyika. J Fish Biol. 76(8):1940–1957.http://dx.doi.org/10.1111/j.1095-8649.2010.02620.x [DOI] [PubMed] [Google Scholar]
- Lee HJ, Kusche H, Meyer A.. 2012. Handed foraging behaviour in scale-eating cichlid fish: Its potential role in shaping morphological asymmetry. PLoS One 7(9):e44670.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee HJ, Heim V, Meyer A.. 2015. Genetic and environmental effects on the morphological asymmetry in the scale-eating cichlid fish, Perissodus microlepis. Ecol Evol. 5(19):4277–4286.http://dx.doi.org/10.1002/ece3.1691 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee HJ, Heim V, Meyer A.. 2016. Genetic evidence for prevalence of alloparental care in a socially monogamous biparental cichlid fish, Perissdous microlepis, from Lake Tanganyika supports the “selfish shepherd effect” hypothesis. Ecol Evol. 6(9):2843–2853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lein E. 2012. Investigation of the correlation between lateralized foraging behavior and neuro-anatomical brain asymmetry in a scale-eating cichlid fish. BSc Thesis. University of Konstanz, Konstanz, Germany.
- Lein ES, et al. 2007. Genome-wide atlas of gene expression in the adult mouse brain. Nature 445(7124):168–176.http://dx.doi.org/10.1038/nature05453 [DOI] [PubMed] [Google Scholar]
- Li B, Dewey C.. 2011. RSEM: accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinformatics 12:323.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacNeilage PF, Rogers LJ, Vallortigara G.. 2009. Origins of the left and right brain. Sci Am. 301(1):60–67. [DOI] [PubMed] [Google Scholar]
- Manousaki T, et al. 2013. Parsing parallel evolution: ecological divergence and differential gene expression in the adaptive radiations of thick-lipped Midas cichlid fishes from Nicaragua. Mol Ecol. 22(3):650–669.http://dx.doi.org/10.1111/mec.12034 [DOI] [PubMed] [Google Scholar]
- Matthews M, Varga ZM.. 2012. Anesthesia and euthanasia in zebrafish. ILAR J 53(2):192–204.http://dx.doi.org/10.1093/ilar.53.2.192 [DOI] [PubMed] [Google Scholar]
- Meyer A. 1987. Phenotypic plasticity and heterochrony in Cichlasoma managuense (Pisces, Cichlidae) and their implications for speciation in cichlid fishes. Evolution 41(6):1357–1369.http://dx.doi.org/10.1111/j.1558-5646.1987.tb02473.x [DOI] [PubMed] [Google Scholar]
- Muschick M, Barluenga M, Salzburger W, Meyer A.. 2011. Adaptive phenotypic plasticity in the Midas cichlid fish pharyngeal jaw and its relevance in adaptive radiation. BMC Evol Biol. 11:116.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nadler JJ, et al. 2006. Large-scale gene expression differences across brain regions and inbred strains correlate with a behavioral phenotype. Genetics 174(3):1229–1236.http://dx.doi.org/10.1534/genetics.106.061481 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Northcutt RG, Wullimann MF.. 1988. The visual system in teleost fishes: morphological patterns and trends In: Atema J, Fay RR, Popper AN, Tavolga WN, editors. Sensory biology of aquatic animals. New York: Springer; p. 515–552. [Google Scholar]
- Overmier JB, Patten RL.. 1982. Teleost telencephalon and memory for delayed reinforcers. Physiol Psychol. 10(1):74–78.http://dx.doi.org/10.3758/BF03327010 [Google Scholar]
- Palmer AR. 2012. Developmental plasticity and the origin of novel forms: unveiling criptic genetic variation via “use and disuse.” J Exp Zool B Mol Dev Evol. 318(6):466–479. [DOI] [PubMed] [Google Scholar]
- Pollen AA, et al. 2007. Environmental complexity and social organization sculpt the brain in Lake Tanganyikan cichlid fish. Brain Behav Evol. 70(1):21–39.http://dx.doi.org/10.1159/000101067 [DOI] [PubMed] [Google Scholar]
- Raffini F, Fruciano C, Franchini P, Meyer A.. 2016. Towards understanding the genetic basis of mouth asymmetry in the scale-eating cichlid Perissodus microlepis. Mol Ecol. 26(1):77–91. [DOI] [PubMed] [Google Scholar]
- R Core Team (2016). R: A language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria. Available from: https://www.R-project.org/, last accessed October 23, 2017.
- Renn SCP, Aubin-Horth N, Hofmann HA.. 2008. Fish and chips: functional genomics of social plasticity in an African cichlid fish. J Exp Biol. 211(Pt 18):3041–3056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogers LJ, Deng C.. 1999. Light experience and lateralization of the two visual pathways in the chick. Behav Brain Res. 98(2):277–287.http://dx.doi.org/10.1016/S0166-4328(98)00094-1 [DOI] [PubMed] [Google Scholar]
- Rogers LJ, Andrew RJ.. 2002. Comparative vertebrate lateralization. Cambridge: Cambridge University Press. [Google Scholar]
- Salas C, et al. 2006. Neuropsychology of learning and memory in teleost fish. Zebrafish 3(2):157–171.http://dx.doi.org/10.1089/zeb.2006.3.157 [DOI] [PubMed] [Google Scholar]
- Schneider RF, Meyer A.. 2017. How plasticity, genetic assimilation and cryptic genetic variation may contribute to adaptive radiations. Mol Ecol. 26(1):330–350.http://dx.doi.org/10.1111/mec.13880 [DOI] [PubMed] [Google Scholar]
- Sokal RR, Rohlf FJ.. 1995. Biometry: the principles and practice of statistics in biological research. New York: W. H. Freeman. [Google Scholar]
- Sommer I, Ramsey N, Kahn R, Aleman A, Bouma A.. 2001. Handedness, language lateralization and anatomical asymmetry in schizophrenia: meta-analysis. Br J Psychiatry 178(4):344–351.http://dx.doi.org/10.1192/bjp.178.4.344 [DOI] [PubMed] [Google Scholar]
- Stewart TA, Albertson RC.. 2010. Evolution of a unique predatory feeding apparatus: functional anatomy, development and a genetic locus for jaw laterality in Lake Tanganyika scale-eating cichlids. BMC Biol. 8(1):11.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun T, et al. 2005. Early asymmetry of gene transcription in embryonic human left and right cerebral cortex. Science 308(5729):1794–1798.http://dx.doi.org/10.1126/science.1110324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun T, Walsh CA.. 2006. Molecular approaches to brain asymmetry and handedness. Nat Rev Neurosci. 7(8):655–662.http://dx.doi.org/10.1038/nrn1930 [DOI] [PubMed] [Google Scholar]
- Takeuchi Y, Hori M, Oda Y.. 2012. Lateralized kinematics of predation behavior in a Lake Tanganyika scale-eating cichlid fish. PLoS One 7(1):e29272.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takeuchi Y, Hori M, Tada S, Oda Y, Salzburger W.. 2016. Acquisition of lateralized predation behavior associated with development of mouth asymmetry in a Lake Tanganyika scale-eating cichlid fish. PLoS One 11(1):e0147476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tremblay KD, Hoodless PA, Bikoff EK, Robertson EJ.. 2000. Formation of the definitive endoderm in mouse is a Smad2-dependent process. Development 127(14):3079–3090. [DOI] [PubMed] [Google Scholar]
- Vallortigara G, Rogers LJ.. 2005. Survival with an asymmetrical brain: Advantages and disadvantages of cerebral lateralization. Behav Brain Sci. 28(4):575–633. [DOI] [PubMed] [Google Scholar]
- Vallortigara G, Rogers LJ, Bisazza A.. 1999. Possible evolutionary origins of cognitive brain lateralization. Brain Res Rev. 30(2):164–175.http://dx.doi.org/10.1016/S0165-0173(99)00012-0 [DOI] [PubMed] [Google Scholar]
- Vallortigara G, Chiandetti C, Sovrano VA.. 2011. Brain asymmetry (animal). WIREs. Cogn Sci. 2(2):146–157. [DOI] [PubMed] [Google Scholar]
- Van Dooren T, van Goor H, van Putten M.. 2010. Handedness and asymmetry in scale-eating cichlids: antisymmetries of different strength. Evolution 64(7):2159–2165. [DOI] [PubMed] [Google Scholar]
- Van Staaden MJ, Huber R, Kaufman LS, Liem KF.. 1995. Brain evolution in cichlids of the African Great Lakes: brain and body size, general patterns, and evolutionary trends. Zoology 98:165–178. [Google Scholar]
- Vuoksimaa E, Koskenvuo M, Rose RJ, Kaprio J.. 2009. Origins of handedness: a nationwide study of 30,161 adults. Neuropsychologia 47(5):1294–1301.http://dx.doi.org/10.1016/j.neuropsychologia.2009.01.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- White LE, Lucas G, Richard A, Purves D.. 1994. Cerebral asymmetry and handedness. Nature 368(6468):197–198.http://dx.doi.org/10.1038/368197a0 [DOI] [PubMed] [Google Scholar]
- Whitfield CW, Cziko A-M, Robinson GE.. 2003. Gene expression profiles in the brain predict behavior in individual honey bees. Science 302(5643):296–299.http://dx.doi.org/10.1126/science.1086807 [DOI] [PubMed] [Google Scholar]
- Wullimann MF. 1997. The central nervous system In: Evans DH. editor. The physiology of fishes. New York: CRC Press; p. 245–282. [Google Scholar]
- Zapala MA, et al. 2005. Adult mouse brain gene expression patterns bear an embryologic imprint. Proc Natl Acad Sci U S A. 102(29):10357–10362. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.