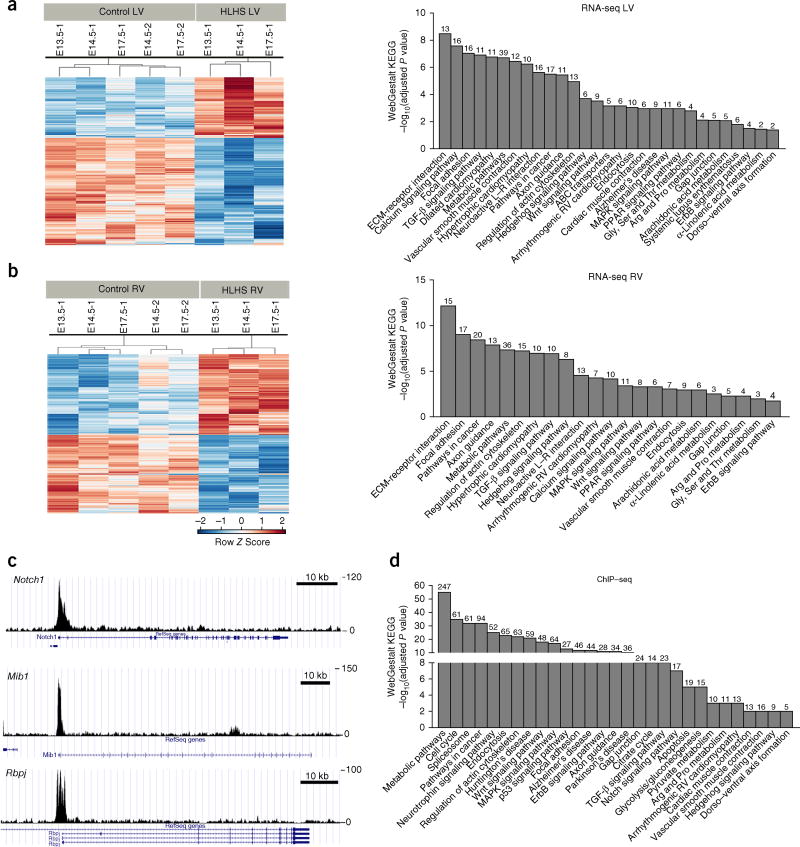
Figure 4.
RNA-seq and Sap130 ChIP–seq analyses show similar pathway enrichment. (a,b) Left, heat maps of RNA-seq conducted with HLHS LV or RV, or littermate-control LV or RV tissues. Shown are log2 c.p.m. values with false discovery rate ≤0.05 and fold change ≥1.5. Right, WebGestalt KEGG pathway enrichment observed for the HLHS LV and HLHS RV RNA-seq. (c) Sap130 ChIP–seq of wild-type mouse whole heart at E12.5, showing Sap130 occupancy over the promoter regions of genes in the Notch signaling pathway (Notch1, Mib1, and Rbpj). Scale bar, 10 kb. (d) WebGestalt KEGG pathway enrichment analysis of Sap130-target genes recovered from Sap130 ChIP–seq. Numbers above bars indicate numbers of genes mapped to each term in the data set.